The regulation of hematopoiesis involves the interaction of specific hematopoietic cytokines with lineage-specific transcription factors, but little is known about how these cytokines might regulate the expression/activity of these different transcription factors. Here we identify the critical signal transduction pathways that mediate the interleukin 3 (IL-3)–induced enhancement of retinoic acid receptor (RAR) transcriptional activity that accompanies the IL-3–mediated commitment of the multipotent, stem cell factor (SCF)–dependent EML cell line to granulocyte/monocyte progenitors. We observe that the addition of IL-3 to EML cells induces activation of the phosphatidylinositol-3 kinase, mitogen-activated protein kinase, and Jak/Stat pathways and that Jak2 activation is the critical “proximal” mediator of the IL-3–induced enhancement of RAR activity. Constitutively active Stat5 constructs enhance both the transcriptional activity of RARs in EML cells and the commitment of these cells to granulocyte/monocyte progenitors, whereas dominant-negative Stat5 constructs inhibit this IL-3–induced enhancement of RAR transcriptional activity. We observe that the retinoic acid response element (RARE) used in our RA responsive reporter harbors overlapping Stat/RAR-binding sites. Moreover, coimmunoprecipitation studies indicate an interaction between Stat5 and RARs that is IL-3 dependent. Thus, Stat5 is an important mediator of the IL-3–induced enhancement of RAR transcriptional activity that accompanies the commitment of immature EML cells to the granulocyte/monocyte lineage. Cytokine-mediated physical and functional interactions between Stat5 and RARs may play critical roles in regulating different stages of hematopoiesis.
Introduction
Defining the molecular events that regulate the development of a multipotent hematopoietic stem cell to a lineage-committed, differentiated progenitor is one of the fundamental goals of experimental hematology. Hematopoietic stem cell development is regulated by specific cytokines acting on hematopoietic precursors of different lineages.1 In addition, different hematopoietic transcription factors also play a critical role in directing the commitment and differentiation of hematopoietic stem cells along a particular lineage.2 Indeed, hematopoiesis involves an intricate functional interaction between these lineage-specific growth factors and these lineage-specific transcription factors, but the molecular basis for how hematopoietic cytokines regulate the expression and activity of these lineage-specific transcription factors remains uncertain.
One family of transcription factors that are important regulators of myeloid differentiation is the retinoic acid receptors (RARs). Retinoic acid (RA) regulates the growth and differentiation of primitive normal myeloid precursors in vitro,3 and knock-out mice deficient in RARα/RARγ display an in vitro block to granulocyte differentiation.4 Moreover, the block in myeloid differentiation displayed by acute promyelocytic leukemia (APL) cells is likely secondary to their characteristic 15;17 chromosome translocation that generates the PML-RARα fusion protein harboring dominant-negative activity against the normal RARα.5-8We have previously observed that transducing a dominant-negative RARα construct into normal mouse bone marrow leads to the generation of continuously proliferating, cytokine-dependent hematopoietic cell lines blocked at distinct stages of myeloid differentiation.9,10One of these cell lines (designated EML) remains relatively immature and multipotent in the presence of stem cell factor (SCF), but with the addition of interleukin 3 (IL-3) these cells commit to the granulocyte/monocyte lineage and generate enhanced numbers of granulocyte-macrophage colony-forming units (CFU-GMs).10We have recently observed that this IL-3–mediated commitment of the multipotent SCF-dependent EML cells to granulocyte/monocyte progenitors is accompanied by enhanced transcriptional activity of RARα.11 Importantly, synthetic antagonists of RARα inhibit this IL-3–mediated granulocyte/monocyte progenitor production by EML cells,11 12 indicating that this enhanced RAR transcriptional activity likely plays a critical role in the IL-3–mediated commitment of the multipotent EML cells to monocyte/granulocyte progenitors. Together these observations indicate that RARs are not only involved in mediating terminal granulocytic differentiation but also may regulate earlier events in myelopoiesis including commitment of an immature precursor to the granulocyte/monocyte lineage.
In the present studies, we defined the specific signal transduction pathways that are involved in this IL-3–mediated up-regulation of RAR transcriptional activity that is associated with the commitment of the multipotent, SCF-dependent EML cells to granulocyte/monocyte progenitors. We compared the signal transduction pathways involved in the SCF-mediated maintenance and proliferation of the multipotent EML cells with those activated by these same cells exposed to IL-3. Although we noted a number of different signal transduction pathways activated by IL-3 in the SCF-dependent EML cells, we observed that Jak2/Stat5 activation plays the critical role both in enhancing RAR transcriptional activity and in stimulating the commitment of the immature, multipotent EML cells to the granulocyte/monocyte lineage. Unexpectedly, we noted overlapping Stat5/RAR-binding sites in the RA responsive elements (RAREs) of a number of different genes. Moreover, we observed an in vivo interaction between Stat5 and RARs that is IL-3 dependent, suggesting that functional cross-talk between Stats and RARs occurs at different stages of hematopoiesis.
Materials and methods
Cell cultures and reagents
Transduction of FLAG-tagged RXR cDNA into BaF3 cells
Using polymerase chain reaction (PCR), a FLAG epitope was introduced at the COOH terminus of the coding region of the human retinoid X receptor α (RXRα) cDNA, and this cDNA was cloned into the LXSN retroviral vector plasmid and packaged into retroviral vector particles as previously described.14 BaF3 cells were infected with these particles followed by G418 selection, and stable cell lines overexpressing the FLAG-tagged RXR were identified using anti-FLAG Western blots.
Expression vector constructs
Expression vectors containing a wild-type Stat5a harboring a COOH-terminus FLAG tag (pRKmStat5acFLAG or Stat5aWT) as well as a dominant-negative Stat5a truncated at amino acid 713 (pRKmStat5a713cFLAG or Stat5aDN) were obtained from Jim Ihle.15 A constitutively active Stat5a mutant (pRKmStat5aHScFLAG or Stat5aHS) was generated from pRKmStat5acFLAG using the site-directed mutagenesis kit (Stratagene, Cedar Creek, TX), by substituting His299 and Ser711 with arginine and phenylalanine, respectively.16 To construct the hybrid GAL-RARα expression vector the complete coding sequence of the human RARα17 was amplified by PCR and cloned into theEcoR1 site of the GALdbd 1-147 expression vector, pSG424.18 An N-terminal deleted GALdbd-RARα fusion construct that includes codons 135 to 462 of RARα (designated GAL-RARΔN) as well as an N- and C-terminal–deleted GALdbd-RARα fusion construct including RARα codons 135 to 403 (designated GAL-RARΔNΔC) were similarly constructed as previously detailed.12 The corresponding p(UAS)5-LUC reporter that is activated by GAL4-RAR in a retinoid-responsive manner has been previously described.12
Antibodies and chemical inhibitors
Rabbit polyclonal antibodies recognizing the phosphorylated p44/42 (Erk1/2) (Thr202/Tyr204), the phosphorylation-independent p44/42, the phosphorylated JNK (Thr183/Tyr185), the phosphorylated p38 mitogen-activated protein (MAP) kinase (Thr180/Tyr182), the phosphorylated Akt (Ser473), the phosphorylation-independent Akt, the phospho-Stat1 (Tyr701), phospho-Stat3 (Tyr705), and phospho-Stat5 (Tyr694) were obtained from Cell Signaling Technology (Beverly, MA). Rabbit antibodies recognizing RARα, RXRα, and the phosphorylation-independent Stat5 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The various chemical inhibitors of the signal transduction pathways including the MEK1/2 inhibitors (PD98059, U0126), the phosphatidylinositol-3 (PI3) kinase inhibitors (wortmannin, Ly294002), and the jak2 (AG490) and jak 3 inhibitors were obtained from Calbiochem (La Jolla, CA). These inhibitors were used at the following concentrations: PD98059, 20 μM; U0126, 10 μM); wortmannin, 200 nM; Ly294002, 20 μM; jak2 inhibitor, 400 μM; and jak 3 inhibitor, 200 μM.
Immunoprecipitation and Western blot analysis
Whole-cell protein extracts for Western blots were obtained by briefly sonicating the cell pellets and then boiling the lysates for 5 minutes in sample lysis buffer containing 50 mM Tris (tris(hydroxymethyl)aminomethane; pH 6.8), 0.5% sodium dodecyl sulfate (SDS), 10% glycerol, and 1 mM dithiothreitol (DTT). For immunoprecipitation cells were lysed in lysis buffer containing 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; pH 7.6), 100 mM KCl, 0.1 mM EDTA (ethylenediaminetetraacetic acid), 10% glycerol, 0.1% Nonidet P-40 (NP-40), 1 mM DTT, 2 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin A, 100 μM phenylmethylsulfonyl fluoride, 1.0 mM NaF, and 2 mM sodium vanadate. Lysates were precleared by incubation with normal serum IgG-bound protein A and G beads for 1 hour at 4°C, and then these precleared lysates were immunoprecipitated for 1 to 4 hours at 4°C using specific antibodies and protein A and G Sepharose beads (Sigma, St Louis, MO). Cell extracts (50 μg/lane) or immunocomplexes were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE; 8%) and electroblotted using the Trans Blot Cell (Bio-Rad, Hercules, CA) onto Polyscreen polyvinylidene difluoride (PVDF) transfer membrane (NEN Life Sciences, Boston, MA). Immunoblotting was performed using the indicated primary antibodies (see above) with peroxidase-conjugated goat antirabbit or antimouse antibodies as secondary antibodies (Santa Cruz Biotechnology). Signals were detected with enhanced chemiluminescence using the Supersignal WestPico luminol/enhancer solution (Pierce, Rockford, IL).
Transient transfections and reporter gene assays
All cell lines were transiently transfected by electroporation as previously detailed.12 Electroporation conditions for the EML and the EML plus IL-3 cells were 270 V and 950 μF. The luciferase reporter is the βRARE tk-LUC, which is based on the pBL2CAT2 vector19 with luciferase replacing CAT and which harbors sequences corresponding to the RARE present in the −55 to −33 sequence of the RARβ2 promoter (AGGGTTCACCGAAAGTTCACTCG; the 5–base pair (bp) “spacer” of the DR5 is underlined) cloned into the HindIII site of this vector. As an internal control for transfection efficiency we used the PON838 plasmid, which is a β-galactosidase reporter driven by the β-actin promoter. Twenty-five micrograms of the βRARE tk-LUC reporter and 20 μg of the PON838 control plasmid were generally used for each transfection. Luciferase activity was normalized to β-galactosidase activity, and relative luciferase activity was calculated as the ratio of this normalized value divided by an average baseline value that was arbitrarily set at 1.
EMSA
Nuclear proteins were extracted as previously described.20 Oligonucleotide probes were synthesized and annealed to their complementary oligo by heating to 70°C. Probes were end labeled by T4 polynucleotide kinase using γ-32P-adenosine triphosphate (ATP) and purified on a 15% nondenaturing polyacrylamide gel. The βRARE probe corresponds to the DR5 RARE in the RARβ promoter21,22 and harbors two 6-bp direct repeats separated by a 5-bp “spacer” 5′ AGGGTTCACCGAAAGTTCACTCG 3′ (the direct repeats are underlined). The corresponding mutated oligo, designated βRARE m4, harbors base pair changes in both direct repeats while maintaining the consensus Stat-binding site: 5′ AGatTTCACCGAAAtaagACTCG 3′ (mutated bases in lower case). The oligo harboring the consensus Stat5-binding site (underlined) is 5′ AGATTTCTAGGAATTCAATCC 3′. The chick α-actin promoter fragment is a 135-bpEcoR1-HindIII restriction fragment that binds a ubiquitously expressed nuclear protein.23 Nuclear extract (15 μg) was incubated with radiolabeled probe (10 000 cpm), and electrophoretic mobility shift assay (EMSA) performed as previously detailed.12 In supershift assays nuclear extracts were preincubated with 1 μg anti-Stat1, anti-Stat3, or anti-Stat5 antibody for 10 minutes at 4°C before adding the labeled probe. In competition assays nuclear extracts were preincubated with a 25- or 50-fold molar excess of the designated unlabeled double-stranded oligo.
Assay of EML CFU-GM generation
Following electroporation with the different expression vectors, EML cells were cultured for 6 to 8 hours in liquid suspension at 5 × 105/mL in media containing SCF alone, and the cells were then harvested and washed, and 5 × 104 viable cells were resuspended in 0.7 mL Iscove modified Dulbecco medium (IMDM) supplemented with 0.75 mL 2.2% methylcellulose (Methocult; Stem Cell Technologies, Vancouver, BC, Canada), 5% horse serum, and 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF). Cultures were incubated in 12-well plates (0.7 mL/plate) and GM-CSF–dependent colonies (>20 cells) were counted following 5 to 7 days of incubation in a humidified incubator.
Results
SCF activates the PI3 kinase and MAP kinase pathways in EML cells
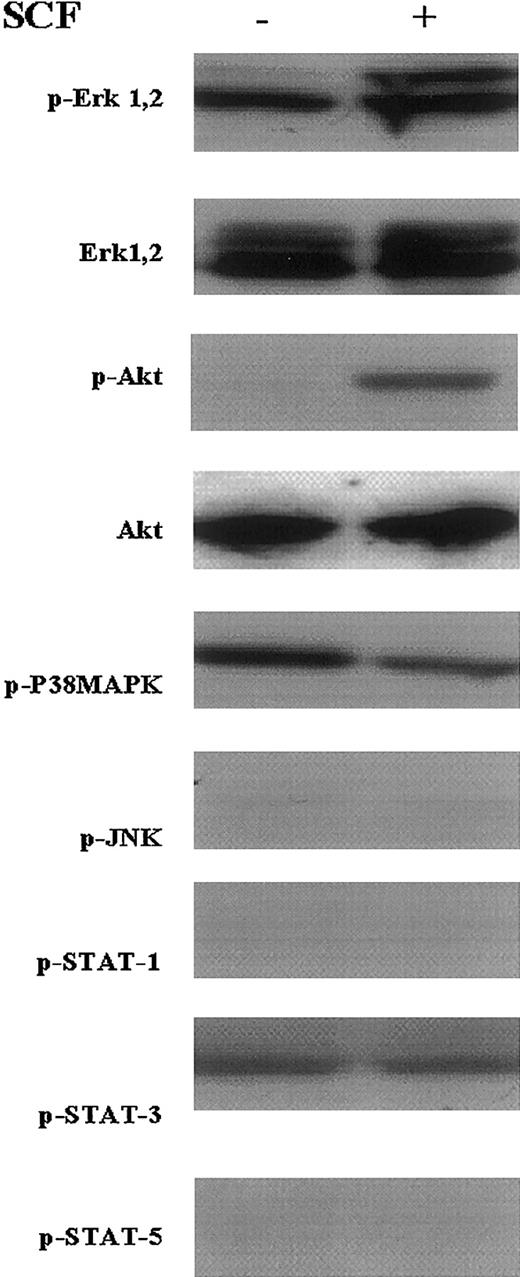
The in vitro proliferation of the multipotent EML cells is absolutely dependent on SCF (kit-ligand), and these cells undergo apoptosis beginning 8 to 12 hours after this cytokine is withdrawn.10 To determine which signal transduction pathways are involved in this SCF-mediated viability/proliferation of the EML cells, we suspended EML cells in SCF-free culture medium and then after 4 hours re-exposed these cells to SCF. Cell lysates obtained following this SCF stimulation were then subjected to Western blot analysis using antibodies detecting activated (phosphorylated) proteins involved in different signal transduction pathways. We observed enhanced phosphorylation of both Erk1/2 (Figure 1, rows 1 and 2) and Akt (Figure 1, rows 3 and 4) following SCF stimulation of the EML cells. We also detected phosphorylated Stat3 and p38 MAP kinase proteins in EML cells in both the presence and absence of SCF (Figure 1, rows 5 and 8). In contrast we observed little if any activation of JNK or of the Stat1 or Stat5 proteins in the EML cells in either the presence or absence of SCF (Figure 1, rows 6, 7, and 9). Thus, in EML cells SCF activates the PI3 kinase (Akt) and MAP kinase (Erk1/2) pathways, whereas Stat3 and p38 MAP kinase appear to be constitutively active in these cells. These observations suggest that SCF maintains EML cell survival/proliferation through activation of the PI3 kinase and MAP kinase pathways.
SCF activates the MAP kinase and PI3 kinase signaling pathways in cultured EML cells.
EML cells were washed and cultured for 4 hours in SCF-deficient media. SCF (50 ng/mL) was then added and Western blots using the indicated antibodies were performed on lysates from cells obtained without (−) and 15 minutes after (+) adding the SCF.
SCF activates the MAP kinase and PI3 kinase signaling pathways in cultured EML cells.
EML cells were washed and cultured for 4 hours in SCF-deficient media. SCF (50 ng/mL) was then added and Western blots using the indicated antibodies were performed on lysates from cells obtained without (−) and 15 minutes after (+) adding the SCF.
IL-3 activates the MAP kinase, PI3 kinase, and Jak/Stat pathways in EML cells
We have previously observed that the addition of IL-3 to the SCF-dependent EML cells markedly stimulates CFU-GM production by these cells.10 12 To determine the specific signal transduction pathways involved in this IL-3–mediated granulocyte/monocyte commitment, we used phosphorylation-specific antibodies to probe Western blots of EML cell lysates obtained at periodic intervals following exposure of the cells to IL-3 (5 ng/mL). In these experiments the IL-3 was added to proliferating EML cells continuously exposed to SCF, and thus this experimental approach assessed the effect of an IL-3 signal superimposed on an SCF signal in the cultured EML cells. The addition of IL-3 enhanced the phosphorylation of both Erk1/2 and of Akt to a degree greater than that observed in the EML cells treated with SCF alone (Figure 2, rows 1-4). We also observed that IL-3 induced the rapid phosphorylation of multiple Stat proteins including Stat1, the higher molecular weight form of Stat3, as well as Stat5 with the phosphorylation of Stat5 appearing particularly prominent (Figure 2, rows 5-7). This phosphorylation occurred within 5 minutes following IL-3 stimulation, peaked before 30 minutes, and then decreased to basal levels after 4 hours (Figure 2). We observed no IL-3–induced activation of either the p38 MAP kinase or JNK pathways in the IL-3–treated cells (not shown). These observations indicate that the addition of IL-3 to the SCF-dependent EML cells activates multiple signal transduction pathways including the MAP kinase, PI3 kinase, and Jak/Stat pathways.
IL-3 activates multiple signal transduction pathways in the SCF-dependent EML cells.
IL-3 (5 ng/mL) was added to EML cells actively proliferating in SCF-containing media. Western blots using the indicated antibodies were then performed on cell lysates harvested at the indicated time points following IL-3 addition.
IL-3 activates multiple signal transduction pathways in the SCF-dependent EML cells.
IL-3 (5 ng/mL) was added to EML cells actively proliferating in SCF-containing media. Western blots using the indicated antibodies were then performed on cell lysates harvested at the indicated time points following IL-3 addition.
Jak2 activation is a critical mediator of Stat, MAP kinase, and PI3 kinase activation in the IL-3–treated EML cells
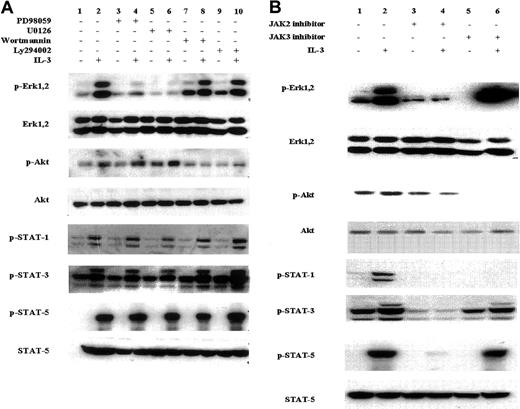
Potentially there may be considerable “cross-talk” between different signal transduction pathways following growth factor/cytokine stimulation. For example, activation of the ras pathway can trigger PI3 kinase activation,24 and the activation of certain receptor-associated Jaks can activate the MAP kinase pathway.25 To determine whether any cross-talk occurs in the Jak/Stat, Erk1/2, and PI3 kinase pathways in the IL-3–induced EML cells, we assessed the effect of specific chemical inhibitors of these different signal transduction pathways. EML cells maintained in SCF were preincubated for 60 minutes with either MEK1/2 inhibitors (PD98059 and U0126), PI3 kinase inhibitors (wortmannin and LY294002), and Jak2 (AG490) or Jak3 inhibitors. Following IL-3 addition (15 minutes), cell lysates were obtained and subjected to Western blot analysis using phosphorylation-specific antibodies. As expected, the IL-3–mediated phosphorylation of the Erk1/2 proteins was blocked by the MEK1/2 inhibitors, and these compounds did not block the IL-3–induced phosphorylation of Akt or of Stat1, Stat3, and Stat5 (Figure 3A, lanes 3-6). Similarly the IL-3–mediated Akt phosphorylation was inhibited by the PI3 kinase inhibitors, but these same compounds exhibited no effect on inhibiting either Stat or Erk1/2 phosphorylation (Figure 3A, lanes 7-10). Thus, in the IL-3–treated EML cells there was little evidence that inhibiting the IL-3 induced activation of either the MAP kinase or PI3 kinase pathways influenced the activity of other signal transduction pathways.
Effect of chemical inhibitors on the IL-3–induced activation of different signal transduction pathways in the SCF-dependent EML cells.
EML cells cultured in SCF-containing media were incubated for 60 minutes in (A) the MEK1/2 inhibitors PD98059 and U0126 (lanes 3-6); the PI3 kinase inhibitors wortmannin and Ly294002 (lanes 7-10); and (B) the Jak2 inhibitor (lanes 3 and 4) and Jak3 inhibitor (lanes 5 and 6). The concentration of these different inhibitors is designated in “Materials and methods.” IL-3 (5 ng/mL) was then added where indicated and Western blots using the indicated antibodies were then performed on cell lysates harvested 15 minutes after the addition of IL-3.
Effect of chemical inhibitors on the IL-3–induced activation of different signal transduction pathways in the SCF-dependent EML cells.
EML cells cultured in SCF-containing media were incubated for 60 minutes in (A) the MEK1/2 inhibitors PD98059 and U0126 (lanes 3-6); the PI3 kinase inhibitors wortmannin and Ly294002 (lanes 7-10); and (B) the Jak2 inhibitor (lanes 3 and 4) and Jak3 inhibitor (lanes 5 and 6). The concentration of these different inhibitors is designated in “Materials and methods.” IL-3 (5 ng/mL) was then added where indicated and Western blots using the indicated antibodies were then performed on cell lysates harvested 15 minutes after the addition of IL-3.
In contrast, exposing the EML cells to the Jak chemical inhibitors clearly influenced multiple signal transduction pathways in the IL-3–treated EML cells. The Jak3 inhibitor inhibits the IL-3–induced Stat1 phosphorylation and partially inhibits the IL-3–induced Stat3 phosphorylation but does not inhibit the induction of Stat5 phosphorylation (Figure 3B, lanes 5 and 6). Curiously, this compound also inhibits Akt phosphorylation but enhances the IL-3–induced Erk1,2 phosphorylation (Figure 3B, columns 5 and 6). In contrast, the Jak2 inhibitor (AG490) exhibited the most “global” inhibitory effects, inhibiting not only Stat1, Stat3, and Stat5 activation in the IL-3–treated cells but also inhibiting the IL-3–induced Erk1,2 and PI3 kinase activation (Figure 3B, columns 3 and 4). This widespread effect of the Jak2 inhibitor in simultaneously inhibiting PI3 kinase, Erk1,2, and Jak/Stat activation strongly suggests that the critical “proximal” event in the IL-3–mediated induction of these multiple signal transduction pathways in EML cells is the activation of Jak2.
The IL-3–induced enhancement of RAR reporter activity is mediated through Jak2
The IL-3–mediated commitment of the SCF-dependent EML cells to the granulocyte/monocyte lineage is associated with the enhanced activity of endogenous RARs in these cells.11 We used the above chemical inhibitors to determine which of the signal transduction pathways activated by IL-3 (Jak/Stat versus Erk1/2 versus PI3 kinase) mediates this enhanced RAR functional activity. IL-3 treatment of the SCF-dependent EML cells enhances the activity of a luciferase reporter construct driven by an RARE11 (Figure 4, column 1). We observed that the MEK1/2 inhibitors (PD98059 and U0126; Figure 4, columns 2 and 3) as well as the PI3 kinase inhibitors (wortmannin and LY294002; Figure 4, columns 4 and 5) did not exhibit any significant effect on this IL-3–mediated enhancement of the RARE luciferase reporter activity. In contrast the Jak2 inhibitor (AG490) markedly inhibited both the IL-3 and the IL-3 plus all-trans-retinoic acid (ATRA)–mediated enhancement of the RARE reporter activity (Figure 4, column 6), whereas the Jak3 inhibitor did not exhibit any inhibition and may actually enhance the RAR activity (Figure 4, column 7). Taken together these observations indicate that Jak2 activation is a critical mediator of the IL-3 enhancement of RAR activity, whereas the PI3 kinase and Erk1/2 pathways exhibit little involvement in this enhanced RAR activity.
The Jak2 inhibitor (AG490) inhibits the IL-3–induced enhancement of RAR transcriptional activity in EML cells.
The SCF-dependent EML cells were incubated overnight (15 hours) in the presence or absence of IL-3 (5 ng/mL) as indicated. Cells were then incubated for 60 minutes in the various chemical inhibitors (respective concentrations are noted in “Materials and methods”). The cells were then electroporated with the βRARE-tk-Luc reporter (25 μg) and the internal control β-galactosidase reporter (20 μg) and after electroporation the cells were maintained in the same inhibitors with or without IL-3 and with or without ATRA treatment (1 μM) as indicated. Five hours later, the cells were harvested and relative luciferase activity was determined on cell extracts as detailed in “Materials and methods.” Results represent the averages and SDs from at least 3 independent experiments.
The Jak2 inhibitor (AG490) inhibits the IL-3–induced enhancement of RAR transcriptional activity in EML cells.
The SCF-dependent EML cells were incubated overnight (15 hours) in the presence or absence of IL-3 (5 ng/mL) as indicated. Cells were then incubated for 60 minutes in the various chemical inhibitors (respective concentrations are noted in “Materials and methods”). The cells were then electroporated with the βRARE-tk-Luc reporter (25 μg) and the internal control β-galactosidase reporter (20 μg) and after electroporation the cells were maintained in the same inhibitors with or without IL-3 and with or without ATRA treatment (1 μM) as indicated. Five hours later, the cells were harvested and relative luciferase activity was determined on cell extracts as detailed in “Materials and methods.” Results represent the averages and SDs from at least 3 independent experiments.
Activated Stat5 is a critical mediator of the IL-3 induction of enhanced RAR reporter activity in EML cells
The above observations indicate that the Jak2/Stat pathway is involved in enhancing the transcriptional activity of the RAR in the IL-3–treated EML cells, but which specific Stat might be the critical mediator of this enhanced activity? We observe that in the IL-3–treated EML cells, the Jak3 inhibitor inhibits Stat1 activation and partially inhibits Stat3 activation but does not inhibit Stat5 activation (Figure 3B, columns 5 and 6), and this compound also does not inhibit the IL-3–induced enhancement of RAR transcriptional activity (Figure 4, column 7). This suggests that Stat5 might be a critical mediator of the IL-3–induced enhancement of RAR activity. To directly test this hypothesis, we determined the effect of both a constitutively active as well as a dominant-negative Stat5 on RAR transcriptional activity in the cultured EML cells. We constructed an expression vector harboring a FLAG-tagged, constitutively active Stat5a mutant (Stat5aHS) in which histamine residue 299 and serine residue 711 were substituted with arginine and phenylalanine, respectively.16 EML cells were cotransfected with the RARE-luciferase reporter together with these different Stat5 expression vectors. Compared with the control (empty) vector as well as the vector harboring the wild-type Stat5a, we observed that transduction of the constitutively active Stat5a into EML cells significantly enhanced RAR transcriptional activity (Figure 5A, column 3).
Activated Stat5 directly mediates the IL-3 induction of enhanced RAR transcriptional activity in EML cells.
(A) Uninduced EML cells were electroporated with the βRARE-tk-Luc reporter (25 μg) together with expression vectors (40 μg of each) for wild-type Stat5 (Stat5aWT) or the constitutively active Stat5 (Stat5aHS). Following overnight incubation, the cells were harvested and relative luciferase activity was determined on cell extracts using the β-galactosidase reporter (20 μg) as an internal control. (B) SCF-dependent EML cells were incubated overnight in the presence or absence of IL-3 (5 ng/mL) as indicated. The cells were then electroporated with the βRARE-tk-Luc reporter (25 μg) together with expression vectors for the indicated Stat5a constructs (40 μg of each). The electroporated cells were then cultured with or without IL-3 for an additional 5 hours, and relative luciferase activity was determined on cell extracts using a β-galactosidase reporter (20 μg) as an internal control. For panels A and B, results represent the averages and SDs from at least 3 independent experiments. (C) SCF-dependent EML cells were stimulated overnight with IL-3. These cells were electroporated with the βRARE-tk-Luc reporter (25 μg) together with the indicated amount of the different Stat5a expression vectors. The cells were then cultured for an additional 5 hours in IL-3 and the relative luciferase activity determined. Results represent the averages and SDs from triplicate samples.
Activated Stat5 directly mediates the IL-3 induction of enhanced RAR transcriptional activity in EML cells.
(A) Uninduced EML cells were electroporated with the βRARE-tk-Luc reporter (25 μg) together with expression vectors (40 μg of each) for wild-type Stat5 (Stat5aWT) or the constitutively active Stat5 (Stat5aHS). Following overnight incubation, the cells were harvested and relative luciferase activity was determined on cell extracts using the β-galactosidase reporter (20 μg) as an internal control. (B) SCF-dependent EML cells were incubated overnight in the presence or absence of IL-3 (5 ng/mL) as indicated. The cells were then electroporated with the βRARE-tk-Luc reporter (25 μg) together with expression vectors for the indicated Stat5a constructs (40 μg of each). The electroporated cells were then cultured with or without IL-3 for an additional 5 hours, and relative luciferase activity was determined on cell extracts using a β-galactosidase reporter (20 μg) as an internal control. For panels A and B, results represent the averages and SDs from at least 3 independent experiments. (C) SCF-dependent EML cells were stimulated overnight with IL-3. These cells were electroporated with the βRARE-tk-Luc reporter (25 μg) together with the indicated amount of the different Stat5a expression vectors. The cells were then cultured for an additional 5 hours in IL-3 and the relative luciferase activity determined. Results represent the averages and SDs from triplicate samples.
We also determined the effect of a dominant-negative Stat5 construct on the IL-3–mediated enhancement of RAR activity in EML cells. In these studies we used a Stat5 expression vector harboring a COOH-terminus truncated Stat5a (Stat5aDN).15 Compared with the wild-type Stat5 vector, this dominant-negative Stat5a (Stat5aDN) significantly inhibits the IL-3–mediated enhancement of RAR activity (Figure 5B). This reduced reporter activity likely is secondary to inhibition of endogenous Stat5 because cotransfection of the wild-type Stat5 construct restores the reporter activity in a dose-dependent manner (Figure 5C). Together these observations using both constitutively active and dominant-negative Stat5 constructs indicate that the activated Stat5 is a critical mediator of the IL-3–induced enhancement of RAR activity in cultured EML cells.
Activated Stat5 enhances CFU-GM generation in the SCF-dependent EML cells
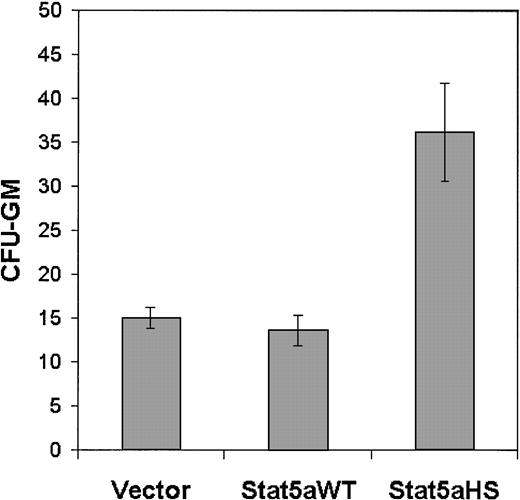
To determine whether Stat5 activation can directly enhance the commitment of the SCF-dependent EML cells to granulocyte/monocyte progenitors, we determined the effect of transducing the constitutively active Stat5 on CFU-GM generation in the cultured EML cells. We observed a significant increase in CFU-GMs in the EML cultures transduced with the activated Stat5 construct (Stat5aHS) compared with the same cells transduced with the control (empty) vector or with the vector harboring the wild-type Stat5a (Figure6). Thus, in the multipotent SCF-dependent EML cells the activation of Stat5 directly enhances the production of granulocyte/macrophage progenitors (CFU-GMs).
Constitutively active Stat5 enhances CFU-GM generation in SCF-dependent EML cells.
EML cells were electroporated with different expression vectors including an empty vector, the wild-type Stat 5 (Stat5aWT), or the constitutively active Stat5 (Stat5aHS; 40 μg of each). Six to 8 hours following electroporation the cells were harvested and assayed for CFU-GM as detailed in “Materials and methods.” Results represent the averages and SDs of 4 independent experiments, and for Stat5aHS,P < .01.
Constitutively active Stat5 enhances CFU-GM generation in SCF-dependent EML cells.
EML cells were electroporated with different expression vectors including an empty vector, the wild-type Stat 5 (Stat5aWT), or the constitutively active Stat5 (Stat5aHS; 40 μg of each). Six to 8 hours following electroporation the cells were harvested and assayed for CFU-GM as detailed in “Materials and methods.” Results represent the averages and SDs of 4 independent experiments, and for Stat5aHS,P < .01.
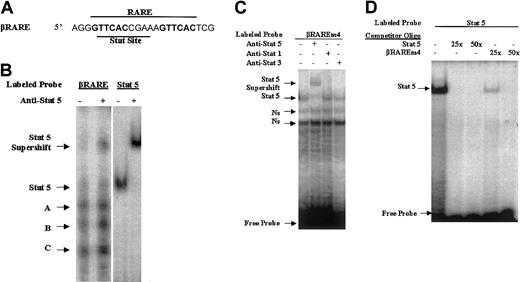
A Stat5-binding site overlaps the RAR-binding site in the RARE within the RARβ promoter
What is the molecular basis for the Stat5-mediated enhancement of RAR activity? The Stats are transcription factors that translocate to the nucleus following phosphorylation by cytokine receptor/Jak complexes,26 and Stat family members generally display binding to the consensus TTC(N)2-4GAA sequence.27 The RARE driving the luciferase reporter in our transfection studies (designated βRARE) is derived from the RARβ promoter (−55 to −33) and is of particular interest because it is likely involved in the autoregulation of RARβ expression in certain cells.21 22 This RARE is a prototype DR5 with a 6-bp repeat separated by a 5-bp “spacer” sequence. Initially we observed that either an oligo harboring a consensus Stat5-binding site or an anti-Stat5 antibody significantly reduced the DNA-binding activity of RAR/RXR to this RARE (data not shown). Close inspection of this sequence reveals a consensus “TTC(N)2-4GAA” Stat-binding site, “TTCACCGAA” that directly overlaps with this DR5 (Figure 7A). To determine whether this consensus Stat-binding sequence observed within the βRARE is indeed a Stat-binding site, we performed an EMSA using nuclear extracts from Stat5-transfected cells incubated with the radiolabeled βRARE oligonucleotide. We observed multiple retarded bands using this oligo likely reflecting the multiple nuclear proteins capable of binding to this sequence (Figure 7B, lane 1). Importantly, the slowest migrating band using the βRARE oligonucleotide migrates at a similar mobility with a single band observed using an oligo harboring a consensus Stat5-binding site alone (Figure 7B, lane 3), and supershift assays with these different oligos using Stat5 antibodies also generate bands of similar mobility (Figure 7B, compare lanes 2 and 4). These EMSAs suggest that the βRARE, in addition to harboring RAR-binding sites, might also harbor sites capable of binding Stat5.
A Stat5-binding site is present in the RARE within the RARβ promoter.
(A) Sequence of the RARE within the RARβ promoter (−55 to −33).21 The two 6-bp direct repeats making up the DR5 element are in bold. (B) Nuclear extracts from BaF3 cells transfected with a Stat5 expression vector were used in an EMSA or an EMSA supershift with the indicated labeled probes and antibody. The Stat5 oligo harbors a consensus Stat5-binding sequence (see “Materials and methods” for this sequence). (C) The βRAREm4 oligo, which harbors mutations within the 6-bp direct repeats while retaining the consensus Stat-binding site (see “Materials and methods” for the exact sequence) served as a probe in EMSAs using BaF3 nuclear extracts and different Stat antibodies as indicated. (D) The labeled Stat5 oligo served as a probe in EMSAs using BaF3 nuclear extracts. Competition reactions were performed with the addition of an excess of different cold oligos as indicated.
A Stat5-binding site is present in the RARE within the RARβ promoter.
(A) Sequence of the RARE within the RARβ promoter (−55 to −33).21 The two 6-bp direct repeats making up the DR5 element are in bold. (B) Nuclear extracts from BaF3 cells transfected with a Stat5 expression vector were used in an EMSA or an EMSA supershift with the indicated labeled probes and antibody. The Stat5 oligo harbors a consensus Stat5-binding sequence (see “Materials and methods” for this sequence). (C) The βRAREm4 oligo, which harbors mutations within the 6-bp direct repeats while retaining the consensus Stat-binding site (see “Materials and methods” for the exact sequence) served as a probe in EMSAs using BaF3 nuclear extracts and different Stat antibodies as indicated. (D) The labeled Stat5 oligo served as a probe in EMSAs using BaF3 nuclear extracts. Competition reactions were performed with the addition of an excess of different cold oligos as indicated.
To confirm a Stat5 interaction with the consensus Stat sequences within the βRARE we performed EMSAs with an oligo harboring mutations that disrupt RAR binding within the βRARE22 while preserving the consensus Stat-binding site. Incubation of this oligo (designated βRAREm4) with nuclear extracts generated a distinct band that was supershifted by Stat5 antibodies but not by antibodies against Stat1 or Stat3 (Figure 7C). Moreover, in competition studies this βRAREm4 oligo competed with the Stat5 binding to the oligo harboring the consensus Stat5-binding site (Figure 7D). Together these observations indicate that the Stat consensus site within the βRARE is indeed capable of binding Stat5.
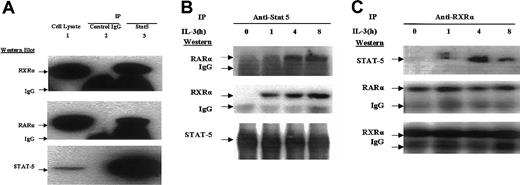
An in vivo interaction between Stat5 and RARs is IL-3 dependent
Our observation that the βRARE oligo harbors overlapping Stat5- and RAR-binding sites (Figure 7) prompted us to perform a coimmunoprecipitation analysis to determine whether Stat5 might interact with the RAR-RXR heterodimer. We have previously encountered difficulty with coimmunoprecipitation studies in EML cells likely secondary to relatively high levels of endogenous protease activity in these particular cells. Therefore, we used the pre-B-cell BaF3 cell line, which proliferates in the presence of exogenous IL-3,13 harbors an activated Stat5,28 and also exhibits Stat5-mediated enhancement of RAR activity (data not shown). We observe that an anti-Stat5 antibody coimmunoprecipitates both RARα and RXRα, whereas control IgG does not (Figure8A). Importantly this coimmunoprecipitation of Stat5 and RAR/RXR occurs only after the cells are exposed to IL-3 (Figure 8B). Similarly, whereas an anti-RXRα antibody coimmunoprecipitates both itself and its RARα partner in IL-3–deprived cells, coimmunoprecipitation of Stat5 with this antibody occurs only in cells treated with IL-3 (Figure 8C). These observations suggest that in at least some cell types there is an in vivo association of Stat5 with components of the RAR-RXR heterodimer but that this association requires the IL-3–mediated activation of Stat5.
Stat5 associates with RXR-RAR in an IL-3–dependent manner.
(A) Cellular lysates (2 mg) of BAF3 cells that had been stably transduced with a FLAG-tagged RXR cDNA (BaF3-RXR cells) were immunoprecipitated with either Stat5 antibody (lane 3) or control IgG (lane 2). The immunoprecipitates, as well as total cell lysate (50 μg; lane 1) were then subjected to Western blot analysis with the indicated antibodies. (B,C) BaF3-RXR cells were deprived of IL-3 for 24 hours and following the addition of IL-3 (5 ng/mL), cell extracts were harvested at the indicated time points. These extracts (1 mg) were immunoprecipitated with antibodies to (B) Stat 5 and (C) RXRα, and the immunoprecipitates were subjected to Western blot analysis with the indicated antibodies.
Stat5 associates with RXR-RAR in an IL-3–dependent manner.
(A) Cellular lysates (2 mg) of BAF3 cells that had been stably transduced with a FLAG-tagged RXR cDNA (BaF3-RXR cells) were immunoprecipitated with either Stat5 antibody (lane 3) or control IgG (lane 2). The immunoprecipitates, as well as total cell lysate (50 μg; lane 1) were then subjected to Western blot analysis with the indicated antibodies. (B,C) BaF3-RXR cells were deprived of IL-3 for 24 hours and following the addition of IL-3 (5 ng/mL), cell extracts were harvested at the indicated time points. These extracts (1 mg) were immunoprecipitated with antibodies to (B) Stat 5 and (C) RXRα, and the immunoprecipitates were subjected to Western blot analysis with the indicated antibodies.
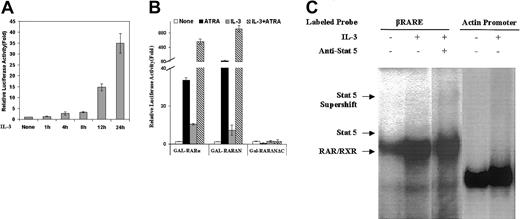
The IL-3–mediated enhancement of βRARE activity is not an immediate early response to Stat5 activation
The observation that the RARE of the RARβ promoter harbors a Stat5-binding site that overlaps with the DR5 (Figure 7A) suggests that the cytokine-activated Stat5 might act in an “immediate early” fashion to directly bind to and activate this promoter element and perhaps displace RAR/RXR binding to the DR5. However, a number of our observations suggest that this is not the case. First, although Stat5 activation occurs very quickly following IL-3 treatment of EML cells (Figure 2, row 7), we observe that the IL-3–induced enhancement of RAR activity requires relatively prolonged exposure to IL-3 (12-24 hours; Figure 9A). Moreover, we note that IL-3 activates an RA-responsive GAL-RARα hybrid on a reporter harboring GAL binding sites (p(UAS)5-LUC; Figure 9B), an experimental model that does not involve any Stat5-binding sites. In addition, we observe in gel shift assays using BaF3 nuclear extracts that IL-3 treatment appears to enhance binding of RAR/RXR to the βRARE (Figure9C). Finally, we note that the βRARE reporter activity is markedly enhanced by ATRA in the IL-3–stimulated EML cells (Figure 4) indicating that the RARs are playing an active role in the IL-3–induced enhancement of this promoter (Figure 4). Taken together these observations suggest that the activation of the βRARE reporter that we have observed in the IL-3–treated cells likely does not result from direct Stat5 activation of this promoter element but rather is somehow mediated through enhanced activity of the RARs.
Stat5 is likely not an immediate early activator of the βRARE.
(A) SCF-dependent EML cells were incubated with IL-3 for the indicated period of time, and the cells were then electroporated with the βRARE-tk-Luc reporter (25 μg). Five hours later, the cells were harvested and relative luciferase activity was determined on cell extracts using a β-galactosidase reporter (20 μg) as an internal control. (B) EML cells as well as EML cells treated overnight with IL-3 as indicated were electroporated with 25 μg of the p(UAS)5-LUC reporter together with 25 μg of each of the indicated plasmids and then cultured in the presence or absence of ATRA (1 μM) as indicated. After 5 hours the relative luciferase activity was determined on cell extracts using a β-galactosidase reporter (20 μg) as an internal control. For panels A and B, results represent the averages and SDs of triplicate samples. As detailed in “Materials and methods” the GAL-RARα and GAL-RARΔN vectors harbor the full-length and N-terminal–deleted RARα cDNA, respectively, whereas the GAL-RARΔNΔC vector also has a deleted C-terminal domain, which renders it unresponsive to ATRA stimulation. (C) BaF3-RXR cells were cultured in IL-3–deficient media for 24 hours. Nuclear extracts from these cells as well as from these same cells stimulated for 1 hour with IL-3 as indicated were used in an EMSA and EMSA supershift with the indicated oligonucleotides and antibody. The actin promoter fragment binds to a ubiquitously expressed nuclear protein23 and thus serves as a control for the amount of nuclear extract used in the EMSA.
Stat5 is likely not an immediate early activator of the βRARE.
(A) SCF-dependent EML cells were incubated with IL-3 for the indicated period of time, and the cells were then electroporated with the βRARE-tk-Luc reporter (25 μg). Five hours later, the cells were harvested and relative luciferase activity was determined on cell extracts using a β-galactosidase reporter (20 μg) as an internal control. (B) EML cells as well as EML cells treated overnight with IL-3 as indicated were electroporated with 25 μg of the p(UAS)5-LUC reporter together with 25 μg of each of the indicated plasmids and then cultured in the presence or absence of ATRA (1 μM) as indicated. After 5 hours the relative luciferase activity was determined on cell extracts using a β-galactosidase reporter (20 μg) as an internal control. For panels A and B, results represent the averages and SDs of triplicate samples. As detailed in “Materials and methods” the GAL-RARα and GAL-RARΔN vectors harbor the full-length and N-terminal–deleted RARα cDNA, respectively, whereas the GAL-RARΔNΔC vector also has a deleted C-terminal domain, which renders it unresponsive to ATRA stimulation. (C) BaF3-RXR cells were cultured in IL-3–deficient media for 24 hours. Nuclear extracts from these cells as well as from these same cells stimulated for 1 hour with IL-3 as indicated were used in an EMSA and EMSA supershift with the indicated oligonucleotides and antibody. The actin promoter fragment binds to a ubiquitously expressed nuclear protein23 and thus serves as a control for the amount of nuclear extract used in the EMSA.
Discussion
Little is known about how specific cytokines might regulate the activity of hematopoietic transcription factors because few experimental models are available to directly approach this question. EML cells, which were derived from normal mouse bone marrow,10 provide a unique in vitro model to address this question because under the influence of SCF these cells retain an immature, multipotent phenotype but can commit to different hematopoietic lineages with the addition of particular cytokines including IL-3, GM-CSF, erythropoietin, and IL-7.10 RARs are important regulators of myeloid differentiation, and we have recently observed that IL-3 enhances the transcriptional activity of RARs in a number of different in vitro models of myeloid differentiation including EML.11 Thus, we were interested in defining the particular signal transduction pathways that mediate this enhanced RAR transcriptional activity in the IL-3–treated EML cells.
We observe that EML exposure to SCF alone is associated with activation of the PI3 kinase and MAP kinase pathways (Figure 1). Similarly, the addition of IL-3 to the SCF-dependent EML cells also activates the PI3 kinase and MAP kinase pathways, but, in addition, the Jak/Stat pathway is activated by IL-3 (Figure 2). Using a battery of specific chemical inhibitors of these different pathways together with constitutively active and dominant-negative constructs, we have identified Stat5 as the critical regulator of the enhanced RAR transcriptional activity that we observe in the IL-3–treated EML cells. This Stat5-mediated enhanced RARα activity appears to be important in the IL-3–induced commitment of the multipotent, SCF-dependent EML cells to the granulocyte/monocyte lineage because specific chemical antagonists of RARα will inhibit the IL-3–induced CFU-GM production by EML cells.11 12
What is the molecular basis for the Stat5-mediated enhancement of RAR transcriptional activity in the IL-3–treated EML cells? The EML cells express a truncated dominant-negative RARα cDNA (RARα403),10 but we have not noted any changes in expression of this transcript in these cells following IL-3 treatment. It is possible that Stat5 may enhance production in EML cells of an endogenous ligand for RAR, but we have observed that conditioned media from IL-3–treated EML cells does not exhibit any activity in enhancing RARE-driven reporter expression (data not shown). Of likely functional significance is our observation that there is a Stat-binding site directly overlapping the RARE within the RARβ promoter (−55 to −33; Figure 7A), which has not previously been noted by other investigators. Moreover, such overlapping sites are not unique to the RARβ RARE, because we observe similar overlapping Stat/RAR sites within the RARE of the RARα2 promoter29 as well as the RARE that lies 3′ of the HOXA1 locus.30 The coevolution of binding sites for both Stats and RARs on such regulatory elements is surprising and suggests a previously unexplored and likely functionally important interaction and cross-talk between the Stat and RAR families of transcription factors. Because reporters driven by this βRARE have been commonly used in studies assessing RAR transcriptional activity in a wide variety of diverse cell types, such studies should be re-evaluated with respect to the contribution of Stat binding in regulating such reporter activity in different cells. How such overlapping Stat/RAR-binding sites might explain the Stat5-mediated enhancement of the βRARE reporter in IL-3–treated cells is presently unclear. However, it appears unlikely that Stat5 acts in an immediate early manner to directly activate the βRARE reporter by binding to and displacing the RAR/RXR receptors from the shared regulatory element because IL-3 treatment appears to enhance rather than inhibit RAR/RXR binding to the βRARE (Figure 9C). Moreover, we have observed that although the IL-3–mediated activation of Stat5 occurs very rapidly, the βRARE reporter response to IL-3 is relatively delayed (Figure9A). In addition, using hybrid GAL-RAR expression vectors we note that IL-3 can enhance basal and ATRA-mediated receptor activity on reporters that lack Stat5-binding sites (Figure 9B).
In addition to such overlapping binding sites, our observation that Stat5 and RARs display an in vivo association that is IL-3 dependent (Figure 8) provides further evidence of a close physical and functional interaction between Stat and RAR family members. Current models of RAR activation suggest that the addition of ligand (ATRA) results in a distinct conformational change in RXR/RAR leading to the release of corepressors and the recruitment of transcriptional coactivators.31 It is possible that activated Stat5 might directly bind to RAR/RXR and trigger a conformational change in the heterodimer that mimics this action of the normal ligand. Alternatively, it has been noted that Stat family members themselves can recruit transcriptional coactivators such as CBP,32and Stat5 might enhance RAR/RXR activity by recruiting additional coactivators to the RARE. Such models are currently quite speculative, and indeed the molecular components making up the Stat5/RAR complex in the IL-3–treated cells are presently unknown. Nevertheless, given the widespread expression and activity of the Stat and RAR families of transcription factors, the precise nature of this Stat5/RAR interaction and its functional consequences are questions worthy of further study.
In summary, we observe that the IL-3–mediated enhancement of RAR activity that is associated with the commitment of the multipotent, SCF-dependent EML cells to granulocyte/monocyte progenitors is associated with the activation of multiple different signal transduction pathways. However, the Jak2/Stat pathway and in particular the activation of Stat5 appears to play the critical role in enhancing both RAR transcriptional activity and CFU-GM production in the SCF-dependent EML cells. Stat5 and RARs associate in vivo in an IL-3–dependent manner suggesting specific physical and functional interactions that likely occur between these transcription factors in certain cytokine-treated cells.
We thank Jim Ihle for the Stat5 constructs, LeMoyne Mueller and Jutta Fero for excellent technical assistance, Ted Gooley for calculating the P values, and Tony Blau for the BaF3 cells.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2001-12-0374.
Supported by National Institutes of Health grants CA58292 (S.J.C.) and HL54881.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Steven J. Collins, Human Biology Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Seattle, WA 98109; e-mail: scollins@fhcrc.org.