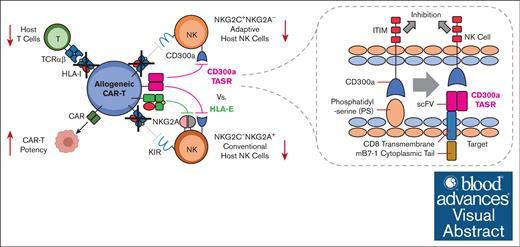
Key Points
An engineered CD300a agonist ligand (CD300a TASR) universally protects HLA-deficient allogeneic T cells from NK cell–mediated rejection.
CD300a TASR enhances chimeric antigen receptor (CAR) T cell efficacy under allogeneic immune pressure.
Visual Abstract
Immunogenicity limits the persistence of off-the-shelf allogeneic cell therapies and transplants. Although ablation of HLA removes most T cell and humoral alloreactivity, no solution has enabled universal protection against the resulting natural killer (NK) cell response. Here, we engineered trans-antigen signaling receptors (TASRs) as a new class of NK inhibitory ligands and discovered CD300a, a previously inaccessible receptor, as a functional target. CD300a TASR outperformed leading alternative strategies in focused screens, including CD47 and HLA-E, and was solely capable of universally protecting allogeneic T cells against a large human cohort (45/45 donors), spanning diverse demographics and NK cell phenotypes. A model allogeneic T-cell therapy coexpressing an anti-CD19 chimeric antigen receptor and CD300a TASR, produced using multiplexed nonviral integration, exhibited enhanced B-cell killing potency under allogeneic immune pressure. CD300 TASR represents a universal solution to NK alloreactivity, broadening the population that could be effectively treated by next-generation allogeneic cell therapies.
Introduction
Off-the-shelf T and chimeric antigen receptor (CAR) T-cell therapies promise to build on the success of its autologous counterparts while providing patients a well-defined, scalable, and cost-efficient drug product that can be administered on demand.1 However, host immune rejection is a major obstacle that limits their persistence, efficacy, and redosing potential.2 Although elimination of donor HLA class I (HLA-I) is a common strategy to evade T-cell alloreactivity and HLA-specific anti-donor antibodies, this strategy also unleashes natural killer (NK) alloreactivity owing to loss of inhibitory signaling from killer cell immunoglobulin-like receptors (KIRs) binding to HLA-I that normally restrains the NK cell response.3 Current approaches to reinstate NK cell inhibition rely on the expression of natural cloaking ligands such as HLA-E,4-6 HLA-A,7 HLA-C,8 and CD479 to agonize natural killer group 2A (NKG2A), KIR, and signal regulatory protein α (SIRPα) inhibitory receptors. However, these targets are only expressed on limited subsets of NK cells or upon sustained cytokine stimulation, rendering the ligands ineffective against hosts with low frequencies of the corresponding NK subset.5,10 In this study, we discover and assess a universal solution against NK alloreactivity to enhance the persistence and efficacy of allogeneic T-cell therapies against all hosts.
Methods
T-cell engineering
Human primary T-cell targets are engineered for knockout (KO) by CRISPR/Cas9 and knockin (KI) by nonviral homology-directed repair (HDR) in accordance with previous protocols.11 HLA-A2 alloreactive T (allo-T) cell clones are generated by orthotopic T-cell receptor α and β (TCRαβ) replacement as previously described.12 For messenger RNA (mRNA)–based screening, 1 to 2 million activated T cells with beta-2 microglobulin (B2M) KO were electroporated in 20 μL P3 buffer containing 1.5 μg of mRNA using code CM-137 and rested overnight. Transgene expression was verified by staining with relevant antibody and/or recombinant protein ligand and acquired on flow cytometry. See supplemental Methods for detailed T-cell engineering, culture, and staining protocols.
Allo-T and NK cell challenge assay
Cryopreserved allo-T cells are thawed, stimulated with Immunocult CD2/CD3/CD28 activator (STEMCELL 10990) and cultured for 6 to 7 days in Rh10p (RPMI, 10% volume-to-volume ratio of human serum, 1× penicillin/streptomycin) supplemented with 40 ng/mL interleukin-2 [IL-2], 10 ng/mL interleukin-7 [IL-7], and 10 ng/mL interleukin-15 [IL-15]). Cryopreserved NK cells are thawed and cultured in Rh10p + 10 ng/mL IL-15 for 3 to 5 days before the start of coculture. For cocultures, 30 000 target T cells are plated with NK and/or allo-T cells at various effector-to-target ratios (E:T ratios) in a 96-well round-bottom plate for 20 hours in R10p (RPMI, 10% v/v fetal bovine serum, 1× penicillin/streptomycin) + 10 ng/mL IL-15 and then subjected to fluorescent barcoding flow cytometry.13
Amine-reactive dyes AF647 (Thermo Fisher Scientific A20006), IF700 (AAT Bioquest 71514), and IR800CW (LI-COR 929-70021) are resuspended in dimethyl sulfoxide and mixed in 8 combinations to make 100× working stocks of 0.2 mg/mL, 1 mg/mL, and 1.5 mg/mL, respectively, and stored in −80°C. The coculture plate was resuspended to 50 μL phosphate buffer saline (PBS) by centrifugation at 500g for 2 minutes to decant supernatant. Fluorescent barcode working stocks were thawed and diluted 1:50 with PBS, and 50 μL of the appropriate barcode combination was immediately transferred to the corresponding wells of the coculture plate using a multichannel pipette and incubated in the dark for 15 minutes at room temperature. Afterwards, 100 μL of R10p was added to each well and incubated for 10 minutes to quench the reaction, followed by 2 washes in staining buffer (PBS, 5 mM EDTA, and 2% v/v fetal bovine serum) by centrifugation at 1200g for 2 minutes. Wells are mixed column-wise and transferred into a new 96-well round-bottom plate, spun 1200g 2 minutes, and then resuspended in the following antibody staining mix in staining buffer: BV421 CD56, BV510 CD3, BV605 HLA-I, 7-AAD, and PE Qbend10. The plate is washed twice in staining buffer by centrifugation at 1200g for 2 minutes and then acquired at equal volumes on flow cytometry.
PBMC challenge assay
Cryopreserved peripheral blood mononuclear cells (PBMCs) are thawed, assessed for phenotype by flow cytometry, and rested overnight in Rh10p + 40 ng/mL IL-2. Notably, 30 000 HDR-edited T cells are plated with PBMCs at various E:T ratios in a 96-well flat-bottom plate for 3 days in R10p + 40 ng/mL IL-2. Cells are transferred to a 96-well round-bottom plate, and T-cell survival is determined by fluorescent barcoding flow cytometry using the protocol outlined above using the following antibody staining mix: PE Qbend10, BV605 HLA-I, BV650 CD56, BV785 CD3, and 7-AAD.
Allo-T + NK cell competition assay
HDR-edited T cells are pooled together at 1:1 ratio, seeded at 200 000 per well, and cocultured with allo-T and NK cells at various E:T ratios in a 96-well flat-bottom plate for 20 hours in R10p + 10 ng/mL IL-15; 10 μL of counting beads (BioLegend 424902) are added to each well and transferred to a new 96-well round-bottom plate. The plate is spun 500g 3 minutes and resuspended into 50 μL of antibody staining mix, consisting of the following reagents in staining buffer: BV421 Qbend10, BV510 CD3, BV605 HLA-I, BV785 CD56, PE 218 Linker, 7-AAD, PE-Cy7 HLA-E, AF647 G4S Linker, and APC-Cy7 CD47. After staining, cells were washed twice with FACS buffer and acquired on flow cytometry at equal volumes per well.
B-cell depletion assay
Cryopreserved PBMCs are thawed and cultured in Rh10p + 40 ng/mL IL-2 for 3 days. PBMCs are seeded at 250 000 per well and cocultured with CAR T cells at various CAR T cell to PBMC ratios in a 96-well flat-bottom plate for 3 days in R10p + 40 ng/mL IL-2. Cells are transferred to a 96-well round-bottom plate spun by centrifugation at 500g for 3 minutes into the following antibody staining mix in 30 μL of staining buffer: BV421 CD56, BV510 CD3, PE CD19, AF647 G4S Linker, 7-AAD are first added, and then 20 μL of an antibody-based fluorescent barcoding mix is added. Barcode staining mix consists of a combination of anti-CD45 antibodies on the BV711, BV785, PE-Cy7, and APC-Cy7 channels suspended in staining buffer that are unique to each row. The plate is washed twice in FACS buffer by centrifugation at 500g for 3 minutes. Wells are mixed column-wise and transferred into a 96-well deep well plate and then acquired at equal volumes on flow cytometry.
All primary cells are obtained from commercial sources from deidentified donors in adherence with the relevant institutional review board and with a written informed consent.
Results
Modeling cellular rejection of allogeneic T cells
We first developed an ex-vivo model of cellular rejection by measuring the survival of human allogeneic T-cell targets cocultured with human primary cell effectors using a modified fluorescently barcoded flow cytometry readout (supplemental Figure 1A).13 This method enabled sensitive and high-throughput measurement of target T-cell survival by direct counting across multiple E:T ratios with high reproducibility in the value of half maximal inhibitory concentration (IC50) (supplemental Figure 1B-C). Analogous to dose-response curves, we define the IC50 value as the E:T ratio corresponding to 50% of target T-cell survival, with higher values indicating enhanced T-cell survival ability. To confirm that disruption of HLA-I eliminates T-cell alloreactivity, we cocultured target T cells from HLA-A2+ serotyped donors with HLA-A2− effector alloreactive T cells engineered to express a known HLA-A2 reactive TCR (Figure 1A). Target T cells were rejected in a dose-dependent manner but were rescued by disruption of HLA-I expression via CRISPR/Cas9-mediated KO of the B2M gene (Figure 1A-B). However, B2M KO sensitized target T cells to NK allorecognition, and neither HLA-I+ nor B2M KO target T cells survived when both NK and alloreactive T cells were added as effectors (Figure 1B). Live cell counts of allo-T and NK cell effectors at assay end point were similar when added alone or together across all E:T ratios against both B2M KO and HLA-I+ targets, demonstrating that cytotoxicity was restricted to target cells and negligible between effector cells (supplemental Figure 2A). B2M KO–mediated NK cell alloreactivity was consistent across multiple donor replicates, confirming missing-self mechanism of NK recognition in this context (supplemental Figure 2B).3,5
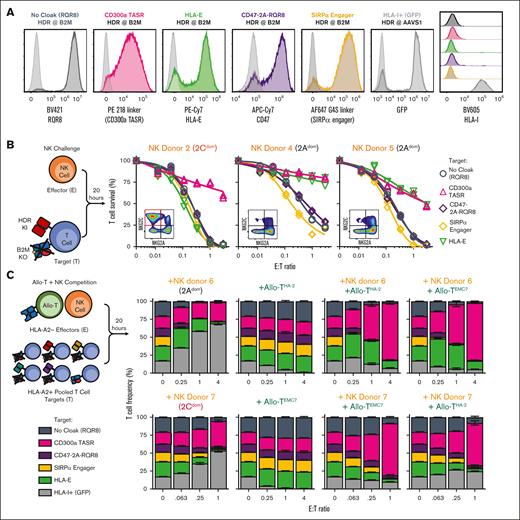
Discovery and optimization of CD300a TASR. (A) Overview of allo–T-cell + NK challenge assay. Survival of HLA-I+ and B2M KO target T cells from an HLA-A2+ donor challenged with NK and allo-T-cell effectors from HLA-A2− donors. Allo-T cells express the AHIII.2 TCRαβ reactive to EMC7 peptide, ALWGFFPVL, presented by HLA-A2. To discern from allo-T cells, B2M KO target T cells were engineered to express RQR8 and HLA-I+ target T cells were engineered to express GFP. (B) HLA-I expression by flow cytometry of HLA-I+ and B2M KO target T cells from panel A. (C) NK challenge assay to screen cloaking strategies on B2M KO T cells. Ligands are expressed by mRNA electroporation (EP), and KO by CRISPR/Cas9. y-axis shows ratio of IC50 value of the indicated cloaked T cell over uncloaked negative control. IC50 represents E:T ratio corresponding to 50% target T-cell survival, derived from one 6-7-point curve of T-cell survival at various E:T ratios challenged with 1 NK cell donor, with higher values indicating enhanced persistence. N = 1 technical replicate per condition. (D) Annotation of TASR structure. (E-G) Effect of hinge length on function of CD300a TASR. (E) Design of CD300a TASR variants with different hinge domains with the indicated amino acid lengths and their expression level after mRNA EP into B2M KO T cells. (F) NK challenge assay expressing the indicated CD300a TASR variant from panel E. N = 2 technical replicate curves per condition. (G) Correlation of NK protection from panel F as defined by IC50 value with hinge length from panel E. Error bar indicates 95% confidence interval of IC50 value. (H-J) Final optimization of CD300a TASR. (H) CD300a TASR V1 and 2 optimized variants are tested for expression via mRNA EP of B2M KO T cells. (I) NK challenge assay against the indicated CD300a TASR variant from (H). N = 1 technical replicate curve per condition. (J) Optimized CD300a TASR design and mechanism of action. GFP, green fluorescent protein; IgG, immunoglobulin G; ITIM, immunoreceptor tyrosine–based inhibitory motif.
Discovery and optimization of CD300a TASR. (A) Overview of allo–T-cell + NK challenge assay. Survival of HLA-I+ and B2M KO target T cells from an HLA-A2+ donor challenged with NK and allo-T-cell effectors from HLA-A2− donors. Allo-T cells express the AHIII.2 TCRαβ reactive to EMC7 peptide, ALWGFFPVL, presented by HLA-A2. To discern from allo-T cells, B2M KO target T cells were engineered to express RQR8 and HLA-I+ target T cells were engineered to express GFP. (B) HLA-I expression by flow cytometry of HLA-I+ and B2M KO target T cells from panel A. (C) NK challenge assay to screen cloaking strategies on B2M KO T cells. Ligands are expressed by mRNA electroporation (EP), and KO by CRISPR/Cas9. y-axis shows ratio of IC50 value of the indicated cloaked T cell over uncloaked negative control. IC50 represents E:T ratio corresponding to 50% target T-cell survival, derived from one 6-7-point curve of T-cell survival at various E:T ratios challenged with 1 NK cell donor, with higher values indicating enhanced persistence. N = 1 technical replicate per condition. (D) Annotation of TASR structure. (E-G) Effect of hinge length on function of CD300a TASR. (E) Design of CD300a TASR variants with different hinge domains with the indicated amino acid lengths and their expression level after mRNA EP into B2M KO T cells. (F) NK challenge assay expressing the indicated CD300a TASR variant from panel E. N = 2 technical replicate curves per condition. (G) Correlation of NK protection from panel F as defined by IC50 value with hinge length from panel E. Error bar indicates 95% confidence interval of IC50 value. (H-J) Final optimization of CD300a TASR. (H) CD300a TASR V1 and 2 optimized variants are tested for expression via mRNA EP of B2M KO T cells. (I) NK challenge assay against the indicated CD300a TASR variant from (H). N = 1 technical replicate curve per condition. (J) Optimized CD300a TASR design and mechanism of action. GFP, green fluorescent protein; IgG, immunoglobulin G; ITIM, immunoreceptor tyrosine–based inhibitory motif.
Screening and discovery of NKG2A and CD300a TASR
We applied this experimental system to rapidly screen previously published “cloaking” strategies known to inhibit NK cell cytotoxicity (Figure 1C; supplemental Figure 3A). One set of strategies involves the removal of ligands that can agonize activating or adhesion receptors on NK cells.14-17 A second set involves the expression of ligands that agonize inhibitory receptors on NK cells or reduce the effect of cytotoxic granules.5,18-21 We challenged B2M KO T cells engineered with each strategy against human primary NK cells from a single donor at various E:T ratios and measured the fold change in T-cell survival relative to noncloaked negative control using the IC50 value. Genetic ablation of CD48, CD54, CD58, and CD155 via CRISPR/Cas9 did not protect against NK cell challenge. This result was reproducible across 2 T-cell lines per target, each engineered with a separate guide RNA with confirmed knockdown of surface expression. Although all natural NK inhibitory ligands were successfully and uniformly expressed via mRNA electroporation, only the well-studied HLA-E enhanced the survival of B2M KO T cells against NK challenge, in line with previous works.5,6
Constrained by the limited targeting repertoire of natural ligand-based approaches, we hypothesized that surface expression of single-chain antigen-binding fragments (scFVs) on an inert scaffold, herein termed trans-antigen signaling receptors (TASRs), can agonize a wider repertoire of NK inhibitory receptors than is currently possible (Figure 1D). We constructed TASRs using an initial CD8 hinge and transmembrane scaffold using scFV constructs of well-studied antibody clones derived from literature and patents to target inhibitory receptors known to be expressed on NK cells. We screened TASRs via mRNA electroporation of B2M KO primary T cells for expression and functional potency against human primary NK cells derived from 1 donor (supplemental Figure 3A). From this screen, we identified a NKG2A and a CD300a-binding TASR that functioned in a dose-dependent manner (Figure 1C; supplemental Figure 3A-C). NKG2A TASR agonizes the same target as HLA-E, whereas CD300a is an unexplored target for this application.
Optimization of CD300a TASR
We set out to optimize the expression and function of CD300a TASR version 1 by testing several mutant constructs using our mRNA screening platform. We discovered a mouse B7-1 transmembrane and cytoplasmic domain that enhanced expression and an inverse correlation between TASR hinge length and the function of CD300a TASR but not NKG2A TASR (Figure 1E-G; supplemental Figures 4 and 5).22 An optimized version 2, containing no hinge but stabilized by the mouse B7-1 cytoplasmic tail and hereafter termed CD300a TASR, exhibited significantly enhanced function while maintaining a fully human extracellular domain (Figure 1H-J).
Validation of CD300a TASR
To model cloaking ligands under constitutive and physiologically relevant levels of expression, we inserted transgenes via CRISPR/Cas9–mediated nonviral HDR of human T cells. Transgenes were integrated into either the B2M locus using the endogenous B2M promoter concomitant with B2M KO or AAVS1 locus using the exogenous elongation factor 1 alpha (EF1α) promoter (Figure 2A; supplemental Figure 6A). CD300a TASR expression was confirmed in both integration approaches by staining with either cognate CD300a protein ligand or an antibody specific to scFV linker sequence (Figure 2B; supplemental Figure 6B). CD300a TASR enhanced T-cell survival in our standard 20-hour challenge assay with IL-15–treated NK cell effectors from multiple donors relative to noncloaked B2M KO T cells (Figure 2C). This result was reproducible with noncytokine-treated NK cells and in long-term 7-day cocultures, with protection on par with HLA-I+ control. CD300a TASR also reduced the secretion of interferon gamma and tumor necrosis factor α release after NK challenge relative to negative control B2M KO T cells and to levels on par with HLA-I+ T-cell positive control (supplemental Figure 6C-D).
Constitutive expression of CD300a TASR robustly protects B2M KO T cells against NK alloreactivity via CD300a agonism. (A) Experimental overview, NK challenge assay with T cells engineered to express cloaking transgene from the B2M locus via nonviral HDR concomitant with B2M KO. (B) TASR and HLA-I expression by flow cytometry of target T cells expressing CD300a TASR or RQR8 negative control transgene from the B2M locus, as well as HLA-I+ control T cells expressing GFP from the AAVS1 locus. CD300a TASR expression evaluated by antibody (left panel) and ligand-based (middle panel) staining. Dashed lines indicate negative control T cells stained with the same marker. (C) NK challenge assay with 3 NK cell donors against target T cells from panels A-B cocultured for 1 and 7 days. NK cells were cultured in 10 ng/mL IL-15, whereas resting NK cells are thawed, rested overnight, and cocultured in the absence of exogenous cytokines. N = 1 to 2 technical replicate curves per condition. (D) 1-day NK challenge assay against target T cells from panels A-B in the presence of 5 μg/mL of anti-CD300a (clone MEM-260), anti-CD300c (clone TX45), or mouse IgG1 isotype control antibody. N = average of 2 technical replicate curves per condition.
Constitutive expression of CD300a TASR robustly protects B2M KO T cells against NK alloreactivity via CD300a agonism. (A) Experimental overview, NK challenge assay with T cells engineered to express cloaking transgene from the B2M locus via nonviral HDR concomitant with B2M KO. (B) TASR and HLA-I expression by flow cytometry of target T cells expressing CD300a TASR or RQR8 negative control transgene from the B2M locus, as well as HLA-I+ control T cells expressing GFP from the AAVS1 locus. CD300a TASR expression evaluated by antibody (left panel) and ligand-based (middle panel) staining. Dashed lines indicate negative control T cells stained with the same marker. (C) NK challenge assay with 3 NK cell donors against target T cells from panels A-B cocultured for 1 and 7 days. NK cells were cultured in 10 ng/mL IL-15, whereas resting NK cells are thawed, rested overnight, and cocultured in the absence of exogenous cytokines. N = 1 to 2 technical replicate curves per condition. (D) 1-day NK challenge assay against target T cells from panels A-B in the presence of 5 μg/mL of anti-CD300a (clone MEM-260), anti-CD300c (clone TX45), or mouse IgG1 isotype control antibody. N = average of 2 technical replicate curves per condition.
To confirm utilization of the CD300a pathway, we performed NK challenge in the presence of blocking antibodies. The addition of anti-CD300a blocking antibody but not isotype control or anti-CD300c, a close paralog of CD300a, completely abrogated the protective effect of CD300a TASR to noncloaked levels (Figure 2D). Anti-CD300a did not affect the survival of noncloaked T cells. Thus, CD300a-mediated inhibition is not functionally used during endogenous T cell and NK cell interactions and the protective effect of CD300a TASR relies on the CD300a signaling axis.
CD300a TASR outperforms alternative strategies
CD300a TASR outperformed the current best-in-class ligands HLA-E and CD47, as well as the recently published SIRPα engager in NK protection.5,6,9,10,23 To provide head-to-head comparisons, we inserted each ligand into the B2M locus of human T cells concomitant with B2M KO and confirmed surface expression (Figure 3A). Challenged with NK cells, CD300a TASR and HLA-E outperformed CD47 and SIRPα engager, whereas CD300a TASR was the only protective ligand against a donor with high frequencies of an NKG2A−NKG2C+ NK cell phenotype, hereby termed 2Cdom (Figure 3B).5 The dominance of CD300a TASR over CD47 was observed under all conditions tested, including when overexpressed via mRNA, inserted into the AAVS1 locus via HDR under EF1α promoter, and challenged with NK cells cultured in high-dose IL-2, which upregulates SIRPα (supplemental Figure 7).10,23 Using mRNA EP, we further determined that CD300a TASR outcompeted TIM3 engager against 4/4 NK donors under comparable levels of expression (supplemental Figure 8).
B2M KO + CD300a TASR outcompetes HLA-E, CD47, and SIRPa engager cloaking ligands and protects against T and NK cells. (A) Phenotype by flow cytometry of engineered T cells expressing the indicated transgenes at the target locus by nonviral HDR, gated single cell lymphocytes. Gray dotted histograms indicate negative control T cells stained with the same markers. CD47 is endogenously expressed on T cells, and so the RQR8 epitope tag was coexpressed using 2A self-cleaving peptide to facilitate purification transgene-expressing cells. (B) NK challenge assay with B2M KO T cells expressing cloaking transgene from the B2M locus from panel A, challenged with 3 NK cell donors. Inset shows the NK cell phenotype by flow cytometry, gated CD3–CD56+. N = 2 technical replicate curves per condition. (C) Competition assay of pooled T-cell targets from panel A cocultured with the indicated allo-T-cell clone and/or NK cell effector donor. Allo-T cells express TCRαβ specific for either minor histocompatibility antigen 2 (HA-2) or EMC7 peptide presented on HLA-A2. Target T cells are serotyped HLA-A2+, and effector cells are HLA-A2−. y-axis shows frequency of the indicated T-cell member after challenge with indicated effector cell. N = 3 technical replicates per condition.
B2M KO + CD300a TASR outcompetes HLA-E, CD47, and SIRPa engager cloaking ligands and protects against T and NK cells. (A) Phenotype by flow cytometry of engineered T cells expressing the indicated transgenes at the target locus by nonviral HDR, gated single cell lymphocytes. Gray dotted histograms indicate negative control T cells stained with the same markers. CD47 is endogenously expressed on T cells, and so the RQR8 epitope tag was coexpressed using 2A self-cleaving peptide to facilitate purification transgene-expressing cells. (B) NK challenge assay with B2M KO T cells expressing cloaking transgene from the B2M locus from panel A, challenged with 3 NK cell donors. Inset shows the NK cell phenotype by flow cytometry, gated CD3–CD56+. N = 2 technical replicate curves per condition. (C) Competition assay of pooled T-cell targets from panel A cocultured with the indicated allo-T-cell clone and/or NK cell effector donor. Allo-T cells express TCRαβ specific for either minor histocompatibility antigen 2 (HA-2) or EMC7 peptide presented on HLA-A2. Target T cells are serotyped HLA-A2+, and effector cells are HLA-A2−. y-axis shows frequency of the indicated T-cell member after challenge with indicated effector cell. N = 3 technical replicates per condition.
As an orthogonal readout, we pooled HDR-edited T cells from the previous experiment along with an HLA-I+ control and challenged with allo-T and NK cells in a competition assay. T cells expressing the indicated ligands were pooled in equal ratios and challenged with NK cells and HLA-A2 alloreactive T cells (Figure 3A,C). The identity and frequency of each T-cell member can be discerned based on antibody staining of the cloaking ligand and measured by conventional multicolor flow cytometry (supplemental Figure 9A-E). CD300a TASR emerged as the dominant survivor when challenged with both NK and allo-T cells (Figure 3C). This result was reproducible across a 2Adom and 2Cdom NK cell donor and 2 alloreactive T-cell clones specific to HA-2 or endoplasmic reticulum membrane protein complex subunit 7 (EMC7) antigen, as well as any combination thereof. In this mixed T-cell model, active T-cell alloreactivity can be observed with both allo–T-cell effectors given that the fraction of HLA-I+ T cells remain significantly lower relative to conditions without allo-T cells. Likewise, active rejection of noncloaked B2M KO T cells can be observed with both NK cell donors in this model relative to conditions without NK cells. Competition with additional NK donors in the absence of HLA-I+ control cells further corroborated dominance of CD300a TASR in 2Cdom NK donors (supplemental Figure 9F).
CD300a TASR is a pan-NK inhibitor
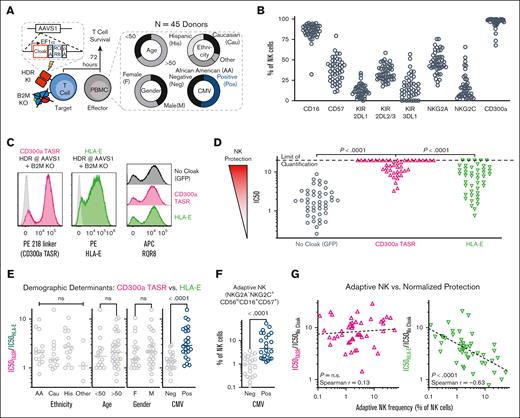
We next assessed CD300a TASR and HLA-E, the most potent alternative, against a large human cohort to assess the heterogeneity of responses across donors and to identify correlates of protection. To model the physiological context of intravenously injected T-cell therapies, we measured the survival of cloaked T cells challenged with PBMCs (Figure 4A; supplemental Figure 10). We selected 45 PBMC donors for diversity in ethnicity, age, gender, and cytomegalovirus (CMV) seropositivity due to their known effects on NK cell function and phenotype (Figure 4A).3,24-26 Notably, although large variations in CD57, KIR, NKG2A, and NKG2C NK marker expression were observed, CD300a was uniformly expressed on NK cells in all donors (Figure 4B; supplemental Figure 11). CD300a TASR and HLA-E were expressed at comparable levels from the AAVS1 locus based on RQR8 expression (Figure 4C).27 Within this cohort, CD300a TASR protected B2M KO T cells against all PBMC donors tested (45/45), indicating universal protection against NK cell reactivity (Figure 4D; supplemental Figure 12). CMV serostatus emerged as the most significant demographic determinant of reduced protection by HLA-E and correlated with the frequency of the well-known NKG2C+NKG2A−CD57+CD16+CD56hi adaptive NK cell subset (Figure 4E-F).28 Although HLA-E lost protection with increasing frequencies of adaptive NK cells, CD300a TASR maintained protection (Figure 4G).
CD300a TASR universally protects against NK cell alloreactivity. (A) Study design and demographic overview of the PBMC donors used. T cells are engineered to coexpress cloaking transgene and RQR8 via 2A self-cleaving peptide under control of EF1α promoter by nonviral HDR. (B) Percentage of NK cells expressing the indicated markers by flow cytometry from the 45 PBMC donors in panel A. (C) Phenotype by flow cytometry of the 3 engineered T-cell targets, gated on live single lymphocytes. Label indicates cloaking transgene. Gray dotted histograms indicate negative control T cells stained with the same markers. (D) Aggregate results as in panel C against 45 PBMC donors. Each data point represents IC50 value of the indicated cloaking ligand against PBMCs from 1 donor. Limit of quantitation in IC50 set to be twice the highest E:T ratio used. Wilcoxon matched pairs signed rank test. (E) Association of PBMC donor demographics from panel A with functional data from panel D, Kruskal-Wallis for ethnicity, Mann-Whitney for other. y-axis represents ratio of IC50 between CD300a TASR and HLA-E cloaking ligand, with 1 indicating equal protection. (F) Adaptive NK cell frequency by CMV serostatus of PBMC donors from panel A. Mann-Whitney U test. (G) Relationship between the adaptive NK cell frequency of the PBMC donor from panel F and functional potency of CD300a TASR (left) and HLA-E (right), normalized to noncloaked control from panel D. Dotted line represents linear fit of log-log transformed data. N = 45 PBMC donors.
CD300a TASR universally protects against NK cell alloreactivity. (A) Study design and demographic overview of the PBMC donors used. T cells are engineered to coexpress cloaking transgene and RQR8 via 2A self-cleaving peptide under control of EF1α promoter by nonviral HDR. (B) Percentage of NK cells expressing the indicated markers by flow cytometry from the 45 PBMC donors in panel A. (C) Phenotype by flow cytometry of the 3 engineered T-cell targets, gated on live single lymphocytes. Label indicates cloaking transgene. Gray dotted histograms indicate negative control T cells stained with the same markers. (D) Aggregate results as in panel C against 45 PBMC donors. Each data point represents IC50 value of the indicated cloaking ligand against PBMCs from 1 donor. Limit of quantitation in IC50 set to be twice the highest E:T ratio used. Wilcoxon matched pairs signed rank test. (E) Association of PBMC donor demographics from panel A with functional data from panel D, Kruskal-Wallis for ethnicity, Mann-Whitney for other. y-axis represents ratio of IC50 between CD300a TASR and HLA-E cloaking ligand, with 1 indicating equal protection. (F) Adaptive NK cell frequency by CMV serostatus of PBMC donors from panel A. Mann-Whitney U test. (G) Relationship between the adaptive NK cell frequency of the PBMC donor from panel F and functional potency of CD300a TASR (left) and HLA-E (right), normalized to noncloaked control from panel D. Dotted line represents linear fit of log-log transformed data. N = 45 PBMC donors.
CD300a TASR enhances anti-CD19 CAR T-cell killing under allogeneic conditions
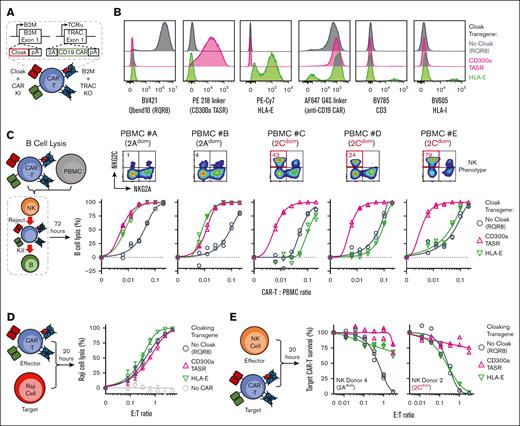
Finally, we integrated recent innovations to produce a model allogeneic CAR T-cell therapy coexpressing a clinically relevant anti-CD19 CAR into the TRAC locus and cloaking ligand into the B2M locus via multiplex nonviral HDR (Figure 5A; supplemental Figure 13A-B).5,11,29 Expression of CD300a TASR, HLA-E, and RQR8 control cloaking ligand in CAR T cells was confirmed (Figure 5B). To model CAR T-cell function under a physiological allogeneic environment, we dosed our engineered CAR T cells into PBMCs and measured killing of CD19-expressing B cells under NK alloreactivity (Figure 5C; supplemental Figure 13C,G). CD300a TASR enhanced B-cell killing potency in all PBMC donors (5/5) and outcompeted HLA-E in all donors with 2Cdom NK phenotype (Figure 5C). This result was not due to intrinsic differences, given that all CAR T cells had similar potency against CD19-expressing Raji cells in the absence of allogeneic pressure (Figure 5D). In addition, TASR-mediated protection against NK cells was maintained in the presence of CAR (Figure 5E).
CD300a TASR enhances CAR T-cell functional potency under allogeneic immune pressure. (A) Nonviral multiplexed targeted integration of human T cells to coexpress cloaking and anti-CD19 CAR transgenes while ablating HLA-I and TCRαβ expression. (B) Phenotype of 3 engineered CAR T cells by flow cytometry expressing the indicated cloaking transgene at B2M locus, gated live single cells. The same anti-CD19 CAR is used for all CAR T cells. (C) B-cell lysis assay for engineered CAR T-cell therapy containing the indicated cloaking transgene against the indicated PBMC donors (bottom row). N = 2 technical replicate curves per condition. Phenotype of NK cells from the respective PBMC donor (top row). (D) Cytotoxicity of indicated cloaked anti-CD19 CAR T cell against CD19-expressing Raji cells. The gray color indicates no CAR control T cells contain CD300a TASR integrated into the B2M loci without anti-CD19 CAR. N = 3 technical replicate curves per condition. (E) NK challenge of cloaked anti-CD19 CAR T cells containing the indicated cloaking transgene with a 2Adom and 2Cdom NK donor. N = 3 technical replicates curves per condition.
CD300a TASR enhances CAR T-cell functional potency under allogeneic immune pressure. (A) Nonviral multiplexed targeted integration of human T cells to coexpress cloaking and anti-CD19 CAR transgenes while ablating HLA-I and TCRαβ expression. (B) Phenotype of 3 engineered CAR T cells by flow cytometry expressing the indicated cloaking transgene at B2M locus, gated live single cells. The same anti-CD19 CAR is used for all CAR T cells. (C) B-cell lysis assay for engineered CAR T-cell therapy containing the indicated cloaking transgene against the indicated PBMC donors (bottom row). N = 2 technical replicate curves per condition. Phenotype of NK cells from the respective PBMC donor (top row). (D) Cytotoxicity of indicated cloaked anti-CD19 CAR T cell against CD19-expressing Raji cells. The gray color indicates no CAR control T cells contain CD300a TASR integrated into the B2M loci without anti-CD19 CAR. N = 3 technical replicate curves per condition. (E) NK challenge of cloaked anti-CD19 CAR T cells containing the indicated cloaking transgene with a 2Adom and 2Cdom NK donor. N = 3 technical replicates curves per condition.
Discussion
Immunological rejection is a major barrier to the persistence and efficacy of allogeneic cell therapies and transplants.30 In this study, we describe the discovery, engineering, and assessment of TASRs as a new modality to inhibit the alloreactive NK cell response. We demonstrate that an optimized CD300a TASR in combination with HLA-I ablation protected against both T and NK cell alloreactivity, exhibited superior function relative to alternative strategies, and possessed broad NK inhibitory potential against a large human cohort. Finally, we show that CD300a TASR can be integrated into a next-generation allogeneic CAR T-cell product via multiplexed, nonviral targeted integration and that the resulting product enhanced B-cell killing potency under allogeneic immune pressure. We expect the B-cell lysis model to be relevant for allogeneic CAR T-cell therapy against systemic lupus erythematosus, where B-cell depletion is the main mechanism of action for dampening the autoreactive antibody response.31
CD300a is a new and unexplored target for NK cloaking. CD300a is a classical inhibitory receptor expressed on all NK cells and signals through immunoreceptor tyrosine–based inhibitory motifs upon binding its endogenous ligands phosphatidylserine and phosphatidylethanolamine (PS/PE) on the surface of apoptotic cells.32-34 Antibody cross-linking studies have also demonstrated Src homology region 2 domain-containing phosphatase 1 (SHP-1) and 2 (SHP-2) recruitment to immunoreceptor tyrosine–based inhibitory motifs to be the primary mechanism of inhibition by CD300a.35-38 Mechanistically, CD300a TASR mirrors the inhibitory effect of anti-CD300a antibody in redirected killing assays with NK effectors against P815 target cells.35 CD300a TASR leaves the host immune system intact and functional, given that noncloaked T cells were still depleted in the presence of CD300a TASR expressing T cells in the competition assay. Thus, CD300a TASR could offer an enhanced safety profile over CAR or antibody-mediated approaches that ablate the host immune system and also leave the endogenous T and NK antitumor response intact.39
The functionality of TASR is determined by both the antigen-binding fragment and the scaffold structure. Despite attempts to reinstate KIR agonism after HLA-I ablation, none of the KIR-targeting TASR clones in our screen demonstrated efficacy. It is possible that our anti-KIR scFVs lack intrinsic agonistic activity or cross reacts with both inhibitory and activating KIRs, which can be difficult to distinguish with TASRs due to high sequence homology between KIR members.40 The beneficial effect of hinge removal for CD300a TASR contrasts with CAR designs, where hinges can enhance CAR T-cell function and signaling.41,42 Given that CD300a TASR is a mimic to the small phospholipids PS/PE, we suspect that hinge removal enhances CD300a agonism by reducing the interaction distance between TASR and CD300a to be comparable with the endogenous interaction of PS/PE and CD300a.
We expect the use of CD300a TASR in allogeneic cell therapies to significantly expand the addressable patient population. By comparing CD300a TASR and HLA-E with a large human cohort, we highlight how NK alloreactivity can exhibit large donor-to-donor variations that should be considered. Although previous work has reported HLA-E’s lack of efficacy against the 2Cdom NK phenotype, our study provides the first comprehensive link among cloaking ligand functional efficacy, NK phenotype, and patient demographics.5 With these data, we were able to robustly show that HLA-E has diminished protection against NK cells isolated from CMV+ individuals whereas CD300a TASR retains function in all hosts independent of demographic determinants. Notably, 50% of CMV+ individuals (12/24) in our cohort harbored substantial adaptive NK cell frequency >5% of total NK cells, which is in line with past estimates and corresponds to the point where HLA-E begins to lose potency in the PBMC challenge assay.28 Given global CMV seroprevalence rates of 43% to 96% by country, we expect CD300a TASR to expand the addressable patient population by 22% to 48% over HLA-E and have the greatest impact in the older individuals aged > 60 years, which have 80%+ CMV seropositivity rate, as is the case with CAR T-cell therapy for most cancer malignancies.43,44
We note several limitations to our study design and scope. First, it remains to be seen whether NKG2A and CD300a TASR can maintain NK protection across a broader range of therapeutic cell types beyond T cells. Second, although CD300a TASR enhanced the persistence and function of an allogeneic CAR T cell produced by multiplexed nonviral integration, additional testing will be required to assess the feasibility of this drug product with respect to genomic integrity, cell phenotype, long-term function, and comparison to other production methods. Finally, additional studies are warranted to investigate the efficacy of TASRs in vivo.
In summary, we have demonstrated that CD300a TASR acts as a universal ligand against NK cell alloreactivity. Its combination with HLA and TCRαβ ablation to prevent T-cell alloreactivity, anti-donor antibodies, and graft-versus-host disease paves the way for fully immunoevasive CAR T cells that are effective for anyone in need of treatment, from cancer to autoimmunity and beyond.
Acknowledgments
The authors thank Chantal Kuhn and Ema Katsumata-Zhang for stimulating discussions and for reviewing the manuscript and Zhenyu Luo for providing the luciferase-expressing Raji cell line.
Authorship
Contribution: S.-Q.Z. conceived and developed the trans-antigen signaling receptor modality; S.-Q.Z. and C.C. conceived and designed the study; S.-Q.Z., F.T., and J.F. designed, performed, and analyzed data for all experiments; S.-Q.Z., K.A., C.C., and G.G.W. supervised the study; and S.-Q.Z. and G.G.W. wrote the manuscript with feedback from all authors.
Conflict of interest disclosure: All authors are former employees of Clade Therapeutics. C.C. is a founder of Clade Therapeutics. A patent application has been filed by Clade Therapeutics for the technology described here.
S-Q.Z. is a current employee of Oncology R&D, AstraZeneca, Waltham, MA. J.F., C.C., and G.G.W. are current employees of Century Therapeutics Inc, Boston, MA.
Correspondence: Shu-Qi Zhang, Clade Therapeutics, Discovery Biology, 201 Brookline Ave, Suite 1002, Boston, MA 02215; email: shuqi.zhang90@gmail.com; and G. Grant Welstead, Clade Therapeutics, Discovery Biology, 201 Brookline Ave, Suite 1002, Boston, MA 02215; email: grant.welstead@centurytx.com.
References
Author notes
C.C. and G.G.W. are joint senior authors.
All data supporting the findings of this study are within the main paper, supplemental Figures, and supplemental Tables. A list of antibody and protein reagents used is in supplemental Table 1. A list of homology-directed repair templates and single-guide RNA sequences are in supplemental Table 2 and 3. A list of messenger RNA–encoding DNA template sequences is in supplemental Table 4.
All other data may be available on request from the corresponding authors Shu-Qi Zhang (shuqi.zhang90@gmail.com) and G. Grant Welstead (grant.welstead@centurytx.com).
The full-text version of this article contains a data supplement.