TO THE EDITOR:
B-cell acute lymphoblastic leukemia (B-ALL) is the most frequent pediatric cancer. This neoplasm of early B cells is a multistep disease featuring diverse genetic alterations.1-3 The prevailing model stipulates that B-ALL occurs in 2 steps: initiation, which often results from a chromosomal translocation, leading to a preleukemic clone, and transformation, which involves acquisition by the preleukemic clone of secondary mutations that promote conversion to overt B-ALL. These mutations often target transcription factors or activate signaling pathways that are involved in B-cell development, resulting in arrested maturation of early B cells.1-3 The events that cause genetic alterations at the initiation step are still obscure. Likewise, despite important advances in the characterization of the secondary mutations, their causal factors, and more generally the molecular mechanisms that underlie the malignant transformation of the preleukemic clone remain poorly understood. Thus, identification of the factors and pathways that influence the preleukemic stage is crucial as the corresponding cells can be the targets of the first malignant transformation hit as well as the source of cells that escape chemotherapy and lead to disease relapse.3
Here, we focused on the importance of interleukin 7 receptor (IL7R) and the recombination activating gene (RAG) 1 and 2 (RAG1/RAG2) complex for the onset of B-ALL. IL7R signaling activates the JAK/STAT, PI3K/AKT, and MAPK/ERK pathways, and regulates multiple fundamental and pathological processes during early B-cell development including proliferation, survival, V(D)J (variable, diversity, and joining) recombination, and leukemia.4 Notably, IL7R was found to protect pre-B cells against the mutagenic activity of the enzyme activation–induced cytidine deaminase (AID).5 The potential involvement of IL7R in B-ALL has been mainly addressed through mutant forms of the IL7R or through the effect of the downstream transcription factor STAT5.6-9 In developing B cells, the RAG complex is absolutely required for V(D)J recombination of immunoglobulin (Ig) genes, but its activity could be oncogenic when it targets non-Ig genes.10,11 For instance, RAG-mediated recombination was found to be the driving mechanism in ETV6-RUNX1–positive B-ALL,10 and in specific instances in cooperation with AID.5,12,13
We have previously generated a mouse model which expresses the human PAX5::ELN oncoprotein (PE) derived from a patient with B-ALL and showed that the model mimics the multistep leukemogenesis process of human B-ALL.14,15 Importantly, the malignant transformation is preceded by 3 months of a preleukemic phase that provides the opportunity for a detailed characterization of abnormal B-cell subpopulations before the onset of leukemia. We addressed the role of IL7R and RAG complex by putting the PAX5::ELN into IL7R- or RAG2-deficient backgrounds (hereafter, PE/IL7R–/– and PE/RAG2–/–, respectively).
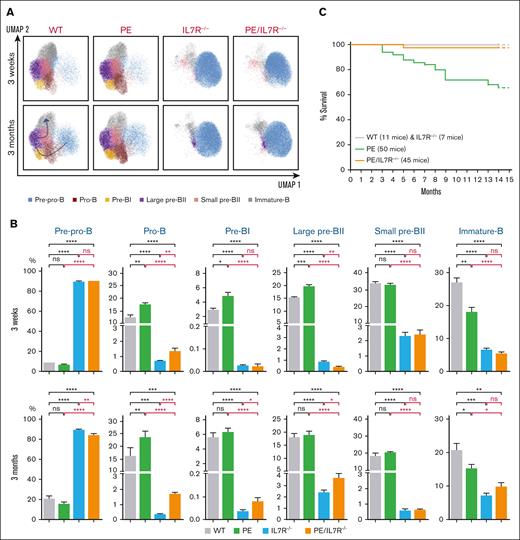
Multiparametric immunophenotyping of medullar B-cell populations from 3-week- and 3-month-old mice revealed a drop of the total medullar B-cell population in PE/IL7R–/– mice associated with a severe, although incomplete, blockade of B-cell development at the pre-pro-B cell stage. This impairment is clearly caused by IL7R-deficiency as PE mice, in contrast to IL7R–/– mice, display no such blockade (Figure 1A-B; supplemental Figures 1, 2A-B, and 3). At subsequent developmental stages, pro-B, pre-BI and large pre-BII PE/IL7R–/– subpopulations tend to increase with age compared with their IL7R–/– counterparts. In contrast, both genotypes display similar percentages of small pre-BII and immature B-cell stages whatever the age of mice (Figure 1A-B). Regardless, from the pro-B stage onward, the percentages of IL7R–/– and PE/IL7R–/– subpopulations remain significantly lower than their PE counterparts (Figure 1A-B).
Effects of PAX5::ELN oncoprotein expression in the absence of IL7R. (A) Immunophenotypic characterization of medullar B-cell populations. UMAP (Uniform Manifold Approximation and Projection) of the clustering analysis of the B-cell subpopulations from 3-week- and 3-month-old mice with the indicated genotypes. Eight surface markers were targeted to identify the subpopulations: B220, CD19, CD117, CD43, IL7R, BP1, CD25, and IgM (supplemental Figure 1 and supplemental Materials and Methods). (B) Percentages of the medullar B-cell subpopulations, from 3-week- and 3-month-old mice with the indicated genotypes. Results are expressed as mean ± SD (GraphPad Prism) and overall differences between values were evaluated by t test with Mann–Whitney U posttest. The difference between means is significant if ∗P value <.05, very significant if ∗∗P value <.01, extremely significant if ∗∗∗P value <.001, or if ∗∗∗∗P value <.0001. (C) Kaplan-Meier survival curves. The genotypes and the numbers of mice are indicated. The follow-up was stopped in the 15th month. Throughout the study, the mutant mice were hemizygous for the human PAX5::ELN–encoding cDNA insertion and homozygous for IL7R-deficiency. IgM, immunoglobulin M; SD, standard deviation; WT, wild type.
Effects of PAX5::ELN oncoprotein expression in the absence of IL7R. (A) Immunophenotypic characterization of medullar B-cell populations. UMAP (Uniform Manifold Approximation and Projection) of the clustering analysis of the B-cell subpopulations from 3-week- and 3-month-old mice with the indicated genotypes. Eight surface markers were targeted to identify the subpopulations: B220, CD19, CD117, CD43, IL7R, BP1, CD25, and IgM (supplemental Figure 1 and supplemental Materials and Methods). (B) Percentages of the medullar B-cell subpopulations, from 3-week- and 3-month-old mice with the indicated genotypes. Results are expressed as mean ± SD (GraphPad Prism) and overall differences between values were evaluated by t test with Mann–Whitney U posttest. The difference between means is significant if ∗P value <.05, very significant if ∗∗P value <.01, extremely significant if ∗∗∗P value <.001, or if ∗∗∗∗P value <.0001. (C) Kaplan-Meier survival curves. The genotypes and the numbers of mice are indicated. The follow-up was stopped in the 15th month. Throughout the study, the mutant mice were hemizygous for the human PAX5::ELN–encoding cDNA insertion and homozygous for IL7R-deficiency. IgM, immunoglobulin M; SD, standard deviation; WT, wild type.
In stark contrast to PE mice, PE/IL7R–/– mice were essentially immune to leukemia for up to 15 months (Figure 1C). This finding clearly indicates that (1) the IL7R is absolutely required for the onset of malignant transformation and suggests that IL7R signaling is an oncogenic relay in B-ALL development induced by PAX5::ELN, and (2) in the absence of IL7R, PAX5::ELN is unable to induce leukemia despite leaky B-cell development. This may be due to the low percentage of pro-B/pre-BI cells that are the target of the oncoprotein, to the preferential expression of PAX5::ELN in CD19+ (ie, post pre-pro-B) subpopulations14,15 or to both, and possibly to a stage-specific requirement for IL7R signaling.
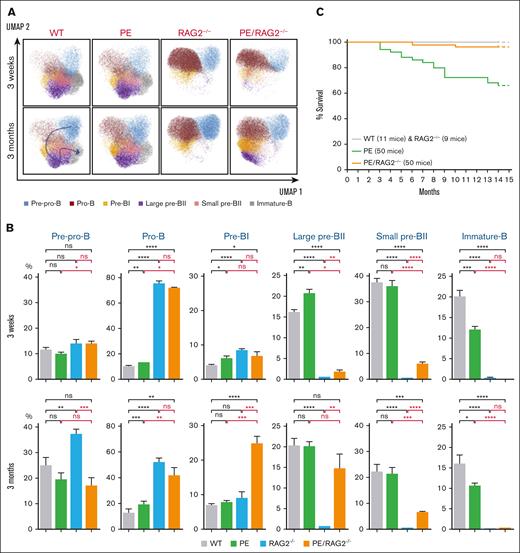
At the genomic level, RAG deficiency suppresses V(D)J recombination, which initiates at the pre-pro-B cell stage. However, at the level of surface markers, RAG-deficiency caused a complete blockade at the later pre-BI/large pre-BII transition (Figure 2A-B; supplemental Figures 1, 2C-D, and 4). Interestingly, PAX5::ELN could overcome this blockade as the subsequent large and small pre-BII stages were readily detected (Figure 2AB) and could account for the relatively higher percentage of total B-cell population in PE/RAG2–/– compared to RAG2–/– 3-month-old mice (supplemental Figure 2CD). In contrast, “phenotypically” immature B-cell population was completely absent in RAG2–/– and PE/RAG2–/– mice (Figure 2A-B).
Effects of PAX5::ELN oncoprotein expression in the absence of the RAG complex. (A) Immunophenotypic characterization of bone marrow B-cell populations. UMAP of the clustering analysis of the B-cell subpopulations from 3-week- and 3-month-old mice with the indicated genotypes. The same combination of markers as in panel B was used. (B) Percentages of the medullar B-cell subpopulations, from 3-week- and 3-month-old mice with the indicated genotypes. Results are expressed as mean ± SD (GraphPad Prism) and overall differences between values were evaluated by t test with Mann–Whitney U posttest. The difference between means is significant if ∗P value <.05, very significant if ∗∗P value <.01, extremely significant if ∗∗∗P value <.001, or if ∗∗∗∗P value <.0001. (C) Kaplan-Meier survival curves. The genotypes and the numbers of mice are indicated. The follow-up and the genetic setting are as in panel A (ie, hemizygous for the PAX5::ELN–encoding cDNA insertion and homozygous for RAG2-deficiency). Note that the follow-up in Figure 1C and panel C was performed at the same time, hence the same cohorts of WT and PE mice were used as controls, the corresponding survival curves have been duplicated for the sake of clarity of the figures.
Effects of PAX5::ELN oncoprotein expression in the absence of the RAG complex. (A) Immunophenotypic characterization of bone marrow B-cell populations. UMAP of the clustering analysis of the B-cell subpopulations from 3-week- and 3-month-old mice with the indicated genotypes. The same combination of markers as in panel B was used. (B) Percentages of the medullar B-cell subpopulations, from 3-week- and 3-month-old mice with the indicated genotypes. Results are expressed as mean ± SD (GraphPad Prism) and overall differences between values were evaluated by t test with Mann–Whitney U posttest. The difference between means is significant if ∗P value <.05, very significant if ∗∗P value <.01, extremely significant if ∗∗∗P value <.001, or if ∗∗∗∗P value <.0001. (C) Kaplan-Meier survival curves. The genotypes and the numbers of mice are indicated. The follow-up and the genetic setting are as in panel A (ie, hemizygous for the PAX5::ELN–encoding cDNA insertion and homozygous for RAG2-deficiency). Note that the follow-up in Figure 1C and panel C was performed at the same time, hence the same cohorts of WT and PE mice were used as controls, the corresponding survival curves have been duplicated for the sake of clarity of the figures.
The Kaplan-Meier curves revealed that the PAX5::ELN oncoprotein could not efficiently induce leukemia in the absence of RAG2 (Figure 2C), indicating that the higher penetrance of leukemia in PE mice is due, at least in part, to the activity of the RAG complex. We conclude that, in our settings, the RAG complex contributes to the onset of leukemia. The requirement for the RAG complex in B-ALL could be through its direct activity on non-Ig genes,10,11 through cooperation with other factors such as AID,5,12,13 indirectly through pre-BCR signaling following a productive V(D)J recombination, or to a combination of these.
We have previously shown that leukemic PE mice accumulated secondary mutations at Ptpn11, Kras, Pax5, and Jak3 genes, recurrently mutated in B-ALL patients.14 We performed targeted sequencing on genomic DNA derived from medullar B cells of 3-months and 6-months old PE/IL7–/– and PE/RAG2–/– mice. Although the recurrent mutations were readily detected in PE mice, none of the 4 genes was mutated in PE/IL7–/– or PE/RAG2–/– mice regardless of their age (supplemental Table 1), suggesting that the lack of secondary mutations is one of the protective mechanisms against B-ALL that result from IL7R- and RAG2-deficiencies.
Overall, this study reveals that the PAX5::ELN by itself is insufficient for an efficient induction of leukemia even in the context of a stalled development and that it requires the cooperation of IL7R and RAG complex, probably in a stage-dependent manner, and at least in part through pathways that ultimately lead to secondary mutations. Whether this requirement relies on the direct effect of IL7R signaling and enzymatic activity of the RAG complex or on the effect of cooperating factors and pathways requires further investigations.
Acknowledgments: The authors thank the Institut de Pharmacologie et de Biologie Structurale (IPBS) animal facility staff and Pénélope Viana from the IPBS cytometry platform for their excellent work, and Naïs Prade and Stéphanie Lagarde from Institut Universitaire du Cancer de Toulouse (IUCT-o) for the targeted sequencing. The authors acknowledge the ANEXPLO/IPBS facility (Genotoul/ANEXPLO), member of the national infrastructure Celphedia, and the TRI IPBS-cytometry (Genotoul-TRI), member of the national infrastructure France-BioImaging, supported by the French National Research Agency (ANR-24-INBS-0005 FBI BIOGEN). This study was supported by the Ligue Contre le Cancer (Ligue Régionale: Comités de l’Ex Région Midi-Pyrénées, N°LS 285415).
Contribution: A.A.K. designed and supervised the study; A.D., E.N., B.G., and A.A.K. designed the methodology; A.D. and E.N. performed the experiments; E.D. supervised targeted sequencing; A.N. performed bioinformatic analyses; A.D. handled the mouse lines; A.A.K. wrote the original draft; A.D., E.N., E.D., A.N., B.G., and A.A.K. reviewed and edited the manuscript; and A.A.K. acquired funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ahmed Amine Khamlichi, Institut de Pharmacologie et de Biologie Structurale, Centre National de la Recherche Scientifique UMR5089, 205 route de Narbonne, 31077 Toulouse cedex, France; email: ahmed.khamlichi@ipbs.fr.
References
Author notes
Original data and/or information on mouse models are available upon reasonable request from the corresponding author, Ahmed Amine Khamlichi (ahmed.khamlichi@ipbs.fr).
The full-text version of this article contains a data supplement.