Key Points
NGS of platelet mtDNA enables in vivo tracking of transfused platelets.
This method can be applied regardless of ethnic differences or multiple transfusions.
Visual Abstract
Tracking transfused platelets is important to evaluate platelet transfusion efficiency. Traditionally, corrected count increments were used; however, quantitative polymerase chain reaction-based methods have recently been developed. As both these methods have some limitations, we developed a new method based on next-generation sequencing (NGS) of platelet mitochondrial DNA (mtDNA). We identified several single nucleotide variant markers by sequencing the entire mtDNA region of platelets, and used NGS to estimate the proportion of each platelet unit. This method was validated using mixed platelets obtained from different donors at various ratios. We confirmed the applicability of this method in patients who received platelet transfusions using pre- and posttransfusion samples from 12 patients with hematological malignancies. The method showed good linearity (r2 >0.99 in the range of mixing ratios from 1:1 to 1:50) in the platelet-mixing experiment. In addition, platelet tracking in patients who received transfusions was feasible using this method. Furthermore, it was possible to track individual platelets in patients who received a single platelet transfusion and in those who received multiple transfusions, including a patient who received 5 platelet transfusions. Hence, this NGS-based platelet-tracking method can be used for patients with various conditions.
Introduction
The evaluation of novel platelet products (such as extended storage, enhanced production automation, application of platelet additive solutions, pathogen-reduction technologies, and other modifications) relies significantly on in vivo measurements of platelet recovery and survival rates after transfusion. Conventionally, the standard technique involved quantifying radioactively labeled platelets.1-4 Furthermore, the Biomedical Excellence for Safer Transfusion Collaborative has proposed a standardized procedure, which involved using indium-111 and chromium-51 to radiolabel platelets.5 However, administering radioactive substances to healthy volunteers is now restricted owing to concerns about radiation exposure.4,6 Moreover, technical limitations4 and a scarcity of radionuclide supplies have limited the use of the radiolabeling method; hence, exploring alternative measuring techniques is necessary.7 Platelet biotinylation is an alternative platelet labeling method, which enables tracking multiple platelet units without the risk of radioactive isotopes.8 However, this method still demands preprocessing of platelets before transfusion. In addition, exposure to biotin may cause the development of antibodies against biotin.8 Detecting differences in HLA phenotypes on the platelet membrane is another approach to discriminating transfused from autologous platelets.9,10 A clear advantage of this tracking method is that special handling or manipulation of the platelet product is not needed, unlike methods using radioactive labeling. However, the need for HLA typing results for both recipients and donors make it difficult to utilize this method. Additionally, this method cannot be used for patients with platelet refractoriness who receive HLA-matched platelet products.
An emerging alternative involves identifying mitochondrial DNA (mtDNA) variants in platelets using quantitative real-time polymerase chain reaction (qRT-PCR)6 or droplet digital polymerase chain reaction (ddPCR).11 This method enables the monitoring of transfused platelet recovery and survival rates without the need for platelet modification. In addition, the variability of mtDNA markers for monitoring platelets in patients was previously discussed by Garritsen et al.12,13 These researchers leveraged genetic variability within the noncoding regions of the mtDNA genome to distinguish platelets from blood samples of various individuals. Notably, 516 base pair substitutions and 151 insertions were identified (ranging from 2-17 variants per donor), by sequencing the noncoding regions of 100 platelet donors.13,14 Subsequently, in other studies, they developed qRT-PCR and ddPCR assays to quantitatively monitor transfused platelets using 5 or 7 single nucleotide variants (SNVs).6,11
In a previous study, mtDNA sequencing by next-generation sequencing (NGS) provided results that were consistent with those of Sanger sequencing,15 providing an attractive alternative to mtDNA sequencing. Hence, this method has the potential to be an efficient and sensitive method for forensic analyses. Additionally, Kim et al demonstrated the feasibility of utilizing SNV-based NGS chimerism assays to monitor engraftment status following allogeneic hematopoietic stem cell transplantation.16 In their investigation, the NGS assay effectively detected occult relapse by distinguishing original malignant cells from donor-derived cells through short tandem repeat analysis. Therefore, the use of NGS to examine mtDNA variants is a promising approach for distinguishing between a patient's platelets and transfused platelets based on the aforementioned studies.
Thus, in this study, we assessed the feasibility of tracking transfused platelets by NGS using both an in vitro platelet mixing study and pre- and post-platelet transfusion samples.
Methods
Participants and study design
We included patients who received single-donor platelet apheresis and had no recent history of platelet transfusion within a week. As a majority of single-donor platelet apheresis are issued to hematology patients, we recruited subjects from this patient group. In total, 12 patients were enrolled, and pre- and posttransfusion peripheral blood samples were collected daily from patients at approximately the same time (between 4 am and 6 am). Sample collection ended when the platelet tracking was no longer available, such as when the patient was discharged or received random donor platelet concentrates (a pool of 6 donors); otherwise, sample collection was conducted for 10 to 12 days following the initial platelet transfusion. This study was reviewed and approved by the institutional review board of the Yonsei University Health System (no. 4-2021-1670). Written informed consent was obtained from all participants enrolled in this study.
In vitro platelet mixing study
We collected small amounts of platelets from 2 ABO-identical platelet concentrates and diluted in phosphate-buffered saline to obtain a platelet suspension of 100 × 109/L. All platelet units were suspended in autologous plasma, and neither irradiation nor pathogen reduction was applied. The 2 diluted platelet concentrates were mixed at ratios of 1:1, 1:5, 1:10, 1:20, and 1:50. Subsequently, we performed NGS-based platelet tracking for each platelet concentrate and the mixed platelet concentrate.
Evaluation of platelet transfusion efficacy
The count of transfused platelets was calculated by multiplying total platelet count by the proportion of transfused platelets obtained from mtDNA sequencing.
DNA extraction, library construction, and sequencing
The platelet component and peripheral blood were filtered using the Acrodisc WBC syringe filter (Pall, Port Washington, NY) for leukoreduction before DNA extraction. This procedure was performed to avoid contaminating DNA originating from white blood cells. Subsequently, mtDNA was extracted from the platelets and peripheral blood using a QIAamp Blood Mini kit (Qiagen, Hilden, Germany), following the manufacturer’s guidelines. In addition, the mtDNA was quantified using a Qubit BR dsDNA kit (Invitrogen, Carlsbad, CA). For mtDNA sequencing, 100 ng of mtDNA was prepared with the Twist Library Preparation EF kit (Twist Bioscience, San Francisco, CA). Furthermore, fragmentation was performed at 32°C for 20 minutes, followed by enzyme inactivation at 65°C for 30 minutes. Target enrichment of mtDNA was performed using a custom-designed enrichment panel following the manufacturer’s instructions (Dxome, Seoul, Republic of Korea). Additionally, the purified libraries were quantified using KAPA Library Quantification kits (Roche Diagnostics, Basel, Switzerland) following the qPCR Quantification Protocol Guide, and qualified using an Agilent Technologies 2200 TapeStation (Agilent Technologies, Santa Clara, CA). Paired-end (2 × 150 bp) sequencing was performed using a NovaSeq platform (Illumina, San Diego, CA).
Bioinformatics analysis
We aligned the mtDNA sequencing data to the mitochondrial reference genome (revised Cambridge Reference Sequence, GenBank NC_012920.1) using BWA-MEM (version 0.7.15). Variant calls were made using GATK Mutect2 (GATK version 4.1.2.0, mitochondria mode) after removing duplicates using Picard. We selected SNVs for further analysis. Furthermore, mtDNA sequencing reads were mapped to the shifted Chr M reference genome to ensure accurate variant calls of the region around the artificial end of circular mtDNA genome (Chr M: 16 024-16 569, and 1-576), and subsequent analyses were conducted as described previously.
Additionally, we excluded any variant with a variant allele frequency (VAF) <1% from the analysis considering the possible errors introduced during the sequencing process. The variants adjacent to well-known error-prone sites in the mtDNA genome were filtered. These sites included Chr M:300-317 and 16 129-16 319. Furthermore, the presence of recipient mitochondria was screened from the sequencing data before platelet transfusion, and variants with a VAF <1% were excluded from further analysis.
Patient data analysis
SNVs of transfused platelets that were different from those of the patient were selected for platelet quantification by comparing the pre- and posttransfusion sequence data. Additionally, donor-specific SNVs were selected from patients who received multiple platelet transfusions. The number of transfused platelets remaining in the patient was calculated by averaging the allele frequencies of unique SNVs from the sequencing data of patient samples collected daily.
Results
Method evaluation using in vitro platelet mixing study
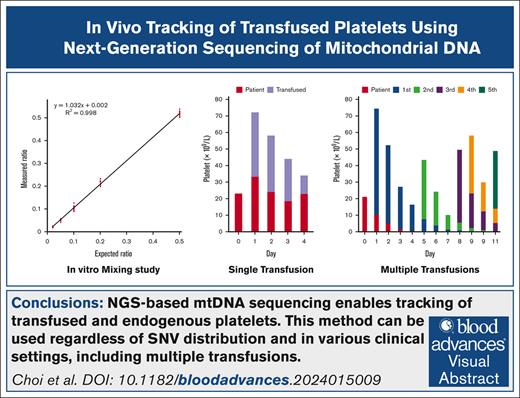
Two sets of ABO-identical platelet concentrates were prepared and serially diluted to a fixed ratio to evaluate mtDNA sequencing-based quantification of platelets. The depth and VAF were constant across all mtDNA regions, except in those adjacent to the error-prone regions. These regions were excluded from the calculation of each platelet proportion for ease of analysis. Even after exclusion, 27 and 18 positions remained as discrimination markers for the 2 sets, respectively. We observed good linearity between the ratio of 1:1 and 1:50 (r2 > 0.99; Figure 1).
In vitro platelet mixing study. Platelet concentrate obtained from donor 1 was mixed with platelet concentrate of donor 2 with a predefined ratio (1:1, 1:5, 1:10, 1:20, and 1:50). Each mixed platelet concentrate was subjected to the NGS-based platelet tracking method, which yielded the ratio of the platelet of donor 1. The ratio of donor 1 platelets at each marker position was plotted (red dots) and used to calculate linearity. The expected and measured ratios of donor 1 platelet are shown.
In vitro platelet mixing study. Platelet concentrate obtained from donor 1 was mixed with platelet concentrate of donor 2 with a predefined ratio (1:1, 1:5, 1:10, 1:20, and 1:50). Each mixed platelet concentrate was subjected to the NGS-based platelet tracking method, which yielded the ratio of the platelet of donor 1. The ratio of donor 1 platelets at each marker position was plotted (red dots) and used to calculate linearity. The expected and measured ratios of donor 1 platelet are shown.
To evaluate whether the storage-dependent mitochondria release of platelet concentrates affects the accuracy of the mtDNA sequencing-based platelet quantification, we performed an additional in vitro mixing experiment. Six pairs of fresh (storage day 3) and old (storage day 5) platelet concentrates were mixed with the ratio of 1:1 and subjected to mtDNA sequencing-based quantification (supplemental Table 1). Across 6 independent mixing sets, the mean ratio of fresh platelets was 50.7% (46.1%-52.9%), while that of old platelets was 49.3% (47.1%-53.9%). These findings suggested that the storage duration of platelet concentrates did not significantly influence the results of the mtDNA sequencing-based quantification method.
Application of platelet tracking using NGS in patients receiving platelet transfusion
We used the NGS-based platelet tracking method for patients with hematologic malignancies who received single-donor platelet apheresis. Peripheral blood samples were collected from each patient before and after transfusion. Among the 12 patients, 1 received a single platelet transfusion, 5 received 2 units of platelets, 2 received 3 units of platelets, and 2 received 4 units of platelets during the evaluation period. Notably, 5 units of platelets were transfused to 2 of the patients. Details of the patients included in this study are described in supplemental Table 2.
The platelet counts of the collected peripheral blood samples ranged from 11 × 109/L to 92 × 109/L. Approximately 1 mL of sample or 1.2 × 107 platelets was sufficient for the extraction of mtDNA and subsequent NGS-based analysis. The yield of platelet units ranged from 2.8 × 1011/unit to 4.9 × 1011/unit with a median value of 3.3 × 1011/unit. The transfused platelets usually remained for 7 days, with the proportion decreasing to <1% thereafter.
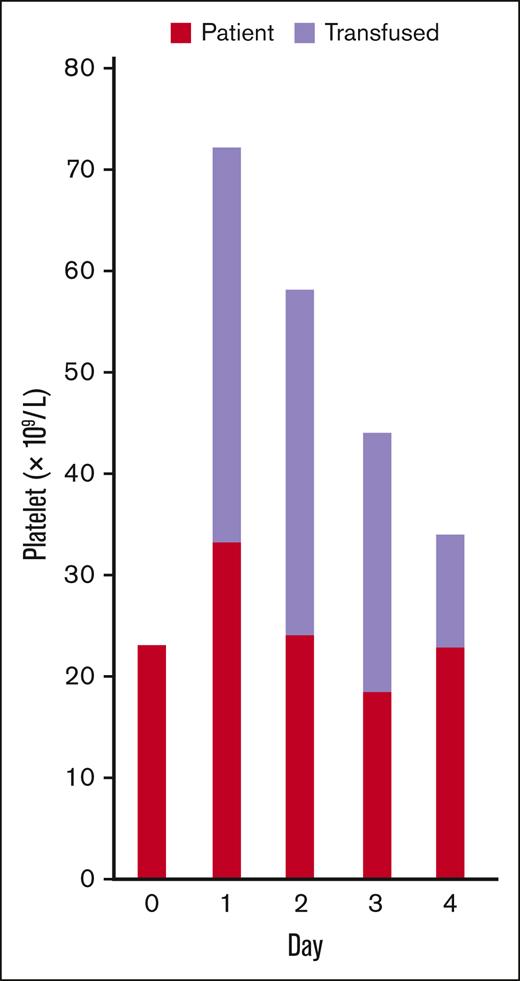
For a patient transfused with only 1 unit of platelets, 21 markers could be used. The analysis results showed that transfused platelet count declined over time, which decreased the total platelet count (Figure 2). The number of endogenous platelets remained within a similar range during the 4-day posttransfusion follow-up period.
Platelet tracking in a patient who received a single platelet transfusion. The counts of patient’s endogenous (red) and transfused (lavender) platelets on each day before and after the transfusion are presented.
Platelet tracking in a patient who received a single platelet transfusion. The counts of patient’s endogenous (red) and transfused (lavender) platelets on each day before and after the transfusion are presented.
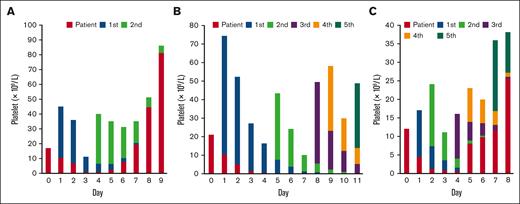
Our methods could be used effectively for tracking transfused platelets, even in patients who received multiple platelet transfusions. We measured each transfused platelet for all patients analyzed, and the results showed either an increase in endogenous platelets (Figure 3A) or a sustained decrease in endogenous platelets (Figure 3B). Additionally, the results showed that our method could track platelets from up to 4 different units, even in a patient who received 5 units of platelet transfusion (Figure 3C). From the sample collected on day 5 after the first platelet transfusion, we effectively discriminated and measured all 4 platelet units transfused within a 5-day period using unique markers specific to the patient and each unit (16, 10, 16, 18, and 9 markers, respectively).
Platelet tracking in patients who received multiple platelet transfusions. The counts of patients’ endogenous (red), and transfused (colored according to the order of transfusion) platelets on each day before and after the transfusion are presented. (A) A patient received 2 platelet transfusions. An increase in patient endogenous platelets was observed from day 5. (B) A patient received 5 platelet transfusions. A sustained decrease in patient endogenous platelets without recovery during the follow-up period was observed. (C) A patient received 5 platelet transfusions within a week. Multiple units of transfused platelets could be discriminated using the method (days 5 and 6).
Platelet tracking in patients who received multiple platelet transfusions. The counts of patients’ endogenous (red), and transfused (colored according to the order of transfusion) platelets on each day before and after the transfusion are presented. (A) A patient received 2 platelet transfusions. An increase in patient endogenous platelets was observed from day 5. (B) A patient received 5 platelet transfusions. A sustained decrease in patient endogenous platelets without recovery during the follow-up period was observed. (C) A patient received 5 platelet transfusions within a week. Multiple units of transfused platelets could be discriminated using the method (days 5 and 6).
Overall, these results reveal the feasibility of NGS-based transfused platelet-tracking methods under various conditions.
Mitochondrial SNV markers for tracking platelets
The human mitochondrial hypervariable region (HVR) contains several SNVs that can be used for individual discrimination. Consistent with previous reports, SNVs in HVR could also be used as markers in our patient cohort. However, some markers frequently used in previous studies, such as those at positions 295, 16 069, and 16 399, could not be used in the present cohort because all patients and donor platelets had the same sequence. Furthermore, position 73 was used as a marker in only 1 patient and position 195 was used in 3 patients. However, position 16 519 was used in 8 patients. Considering the small sample size of our cohort, we searched for the allele frequency (AF) at each position in a large genome database (https://gnomad.broadinstitute.org/). The m.73A>G showed a global AF of 0.7157, which was sufficient as a discrimination marker; however, the AF in East Asians was 0.9973. Moreover, the AFs of the other 3 SNVs that could not be used as markers in our cohort—m.295C>T (global AF, 0.056; East Asian AF, 0), m.16069C>T (global AF, 0.056; East Asian AF, 0), and m.16399A>G (global AF, 0.035; East Asian AF, 0.026)—showed similar results, emphasizing the need to consider ethnic differences when designing platelet-tracking methods using selected mtDNA markers.
Using NGS-based methods, multiple SNVs existing in the HVR and in other areas have been successfully used as markers. The most frequently used marker was m.10398A>G (8 patients), followed by m.152T>C, m.709G>A, m.4883C>T, m.5178C>A, and m.12705C>T, each used in 7 patients. The mtDNA positions of the SNVs used as markers in this study are listed in supplemental Table 3.
Advantages of NGS-based platelet tracking method
We calculated the CCI and compared it with the percentage of transfused platelets for each platelet transfusion. This was calculated based on the NGS-based measured value of transfused platelets. The CCI calculated the day after transfusion (mostly measured 18 hours after transfusion) was >5000 in most of the transfusion episodes. The NGS-based tracking of the percentage of transfused platelets provides an accurate interpretation of the efficacy of platelet transfusion in low CCI cases. Notably, the platelet count decreased despite platelet transfusion in a patient (third transfusion; supplemental Figure 1A), and CCI showed negative values (supplemental Figure 1B). The results of the NGS-based tracking method showed that the extremely low percentage of transfused platelets (4.5%) was responsible for the decrease in posttransfusion platelet count and not the decrease in endogenous platelets (supplemental Figure 1C). In another patient, high body weight resulted in a low CCI (<5000) after the first and third transfusions (supplemental Figure 2A). The percentages of transfused platelets were calculated as 30.5% and 25.1%, respectively, which showed appropriate transfusion efficiency using the NGS-based method (supplemental Figure 2B).
Notably, multiple prior platelet transfusions made it difficult to evaluate the efficiency of platelet transfusions using CCI. A decrease in previously transfused platelets was the cause of the low CCI during the third platelet transfusion in a patient (supplemental Figure 3A-B). In this patient, the percentage of transfused platelets was 24.4% (supplemental Figure 3C), indicating adequate efficiency of the platelet transfusion.
Discussion
Herein, we described a proof-of-concept study for an NGS-based platelet-tracking method. The results of the in vitro mixing study suggest that this method can reliably measure the proportion of platelets in an individual. In addition, this method has been used for patients who received platelet transfusions, demonstrating its feasibility.
Conventionally, the CCI is used to evaluate platelet transfusion efficiency. This method is widely used for its simplicity in calculation, and because it is an objective measure of the effectiveness of platelet transfusion. However, CCI-based evaluations have certain limitations. One of the limitations of this evaluation is that it can only be used correctly when the number of endogenous platelets remains constant. However, in our analysis endogenous platelet counts varied over time in the patients, supporting the use of the platelet-tracking method. In addition, our method is beneficial because patients often require >1 unit of transfusion during hospitalization and the CCI-based methods cannot be used in repetitive transfusion cases.
Previous studies have shown the feasibility of qRT-PCR and ddPCR for in vivo tracking of transfused platelets. These tracking methods have been used and proven useful for tracking transfused platelets in individual patients,6 evaluating pathogen inactivation techniques,18,19 monitoring endogenous platelet recovery trends in patients who have undergone allogeneic stem cell transplantation,20 and assessing the effectiveness of platelet transfusion in patients who have undergone cardiac surgery.11 In these studies, they used 5 to 7 markers located at HVR, with good discriminating power. Hence, the methods used in these studies were robust; however, differences in population frequency in each ethnic background limit the use of the markers in certain populations. Some of the markers cannot be used because of the homogeneity in the population, as shown in our study. However, our results indicate the feasibility of using markers residing outside the HVR. The main advantage of this method is the large set of mtDNA markers that can be used regardless of ethnic differences. In addition, this enables the analysis of patients with multiple platelet transfusions within a short period, in which platelets from different donors coexist in circulation. We demonstrate that this method can be used in a patient who received 5 units of platelet within 8 days, which can discriminate each unit efficiently. Another advantage of the NGS-based platelet tracking method is that the donor typing before transfusion is not mandatory. Since almost all regions of mtDNA are covered, simple serial recipient sampling enables the discrimination of donor platelet by identifying newly introduced variants after each transfusion.
Our method enables the tracking of each transfused platelet, and the results show some notable cases. In one patient, the number of platelets transfused a day after transfusion was unexpectedly high. The increases in posttransfusion platelets may be interpreted as platelet transfusion and an increase in endogenous platelets. However, transfused platelets count measured by NGS-based method was high, whereas endogenous platelets decreased (described in Figure 3B). In contrast, 1 of the 4 transfusions in a patient who received multiple platelet transfusions showed very low recovery and survival on the day after transfusion, and platelets became undetectable 2 days after transfusion (described in supplemental Figure 1). These results indicate that NGS-based platelet tracking can reveal the percentage of transfused platelets in the circulation of the patient, serving as background data for platelet transfusion efficiency and donor selection.
This method has certain limitations. First, the relatively long turnaround time limited the prompt application of the results. However, reducing the overall time required to obtain results can be achieved by applying a recently developed method; for example, using Oxford Nanopore for the analysis may significantly reduce the time to obtain results.21 In addition, the costs of the analysis are much higher than those of qPCR-based platelet tracking and CCI-based measurement of transfused platelets, which may limit the use of the NGS-based tracking method. Second, this method cannot be used if mitochondrial SNV markers are unavailable. Apart from the cases wherein platelet components were donated by maternally related donors, there may be rare cases in which individuals share the same mitochondrial sequences. Third, it would be interesting to apply this method to track HLA-matched platelets transfused in patients with platelet transfusion refractoriness; however, we were unable to analyze these patients. As the recovery and survival of patients with HLA-matched platelets is expected to be much higher than that of patients with HLA-unmatched platelets, future studies should be conducted on these patients. Fourth, we included only patients with hematological malignancies. Patients with various other conditions that require platelet transfusion, such as idiopathic thrombocytopenic purpura, platelet-consuming conditions including liver diseases, and patients undergoing surgery, might benefit from this method, warranting future studies.
In summary, NGS-based mtDNA sequencing enables tracking of transfused and endogenous platelets. This technique can be used regardless of the SNV distribution and can be used in the setting of multiple platelet transfusions. It is expected to be useful for patients in various clinical situations, such as allogeneic stem cell transplant recipients, surgery, and bleeding, and also for evaluating the transfusion efficiency of various platelet products, including stem cell–derived platelets because our method is broadly applicable.
Acknowledgments
This study was supported by a grant of the Manufacturing Human Cell-based Artificial Blood and Platform Technology Development for Transfusion, funded by the Multi-Ministrial Research Project, Republic of Korea (grant number HX23C1706).
Authorship
Contribution: S.J.C. and S.K. conceptualized the study and developed methodology; S.J.C., H.K.K., E.J.S., and S.S.K. curated data and performed analysis; S.K. administered the project and supervised the study; S.J.C. and S.S.K. drafted the manuscript; H.K.K., E.J.S., H. Chung, H. Cho, J.-W.C., and S.K. reviewed and edited the manuscript and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sinyoung Kim, Department of Laboratory Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seoul 03722, Republic of Korea; email: sykim@yuhs.ac.
References
Author notes
S.J.C. and S.S.K. contributed equally to this work.
The data used in this study are available upon reasonable request from the corresponding author Sinyoung Kim (sykim@yuhs.ac).
The full-text version of this article contains a data supplement.