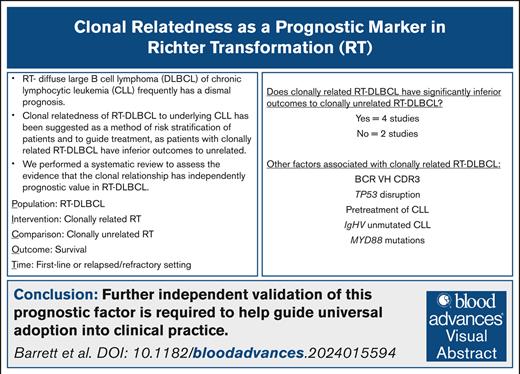
Visual Abstract
The disease shift of chronic lymphocytic leukemia (CLL) to diffuse large B-cell lymphoma (DLBCL), so-called Richter transformation (RT), is a catastrophic clinical event. Rarely, survival can outperform expectations and accurate prognostication for patients may affect therapeutic choices. To date, prognosis has relied on readily available factors such as TP53 disruption, previous CLL treatment status, and performance score. Recently, a shared clonality assessment by immunoglobulin heavy-chain variable region gene (IgHV) sequencing of the CLL and RT has been considered therapeutically relevant, but this is infrequently performed. We performed a systematic review of peer-reviewed manuscripts where outcomes in relation to clonal relatedness and lack thereof (clonally related and clonally unrelated RT-DLBCL) were examined. Fifteen manuscripts that included 336 patients were found, of which 6 compared survival outcomes between the 2 groups in a statistically meaningful way. Two analyses showed no difference in survival outcomes with 4 studies reporting a significantly poorer prognosis with clonally related RT-DLBCL. In 2 of these studies, the baseline characteristics of clonally related and clonally unrelated groups were compared and the clonally related cases were enriched for underlying CLL, which was TP53 disrupted, IgHV unmutated, more heavily pretreated, and exhibiting stereotyped B-cell receptor VH CDR3 and RT-DLBCL, which was MYD88 wild type. We demonstrate that although clonal relatedness of the underlying CLL confers a poorer survival, this is not demonstrated in any study to be independent of other well-described clinical and genomic variables known to influence outcomes in RT-DLBCL. Further independent validation of this prognostic factor is required to help guide universal adoption into clinical practice.
Introduction
Despite significant advances in the treatment of chronic lymphocytic leukemia (CLL) over the last decade, Richter transformation (RT) of underlying CLL into diffuse large B-cell lymphoma (DLBCL) remains a threat to long-term survival of patients with CLL and frequently portends a dismal prognosis.1
The biology of RT-DLBCL remains incompletely understood. Risk factors for RT-DLBCL development include advanced CLL stage at diagnosis, bone marrow involvement, and the need for treatment of CLL (although it can arise in untreated cases or be diagnosed synchronously with CLL).2-4 Genetic aberrations of CLL including unmutated immunoglobulin heavy-chain (IgH) variable region gene (IgHV), NOTCH1, and del(17p) and immunohistochemical features such as increased ZAP70 and CD38 expression also increase the likelihood of RT-DLBCL, with IgH subsets 8 and 3 to 49 conferring particular risk.5,6 RT-DLBCL commonly shares genetic mutations with the underlying CLL clone with frequent demonstration of complex chromosomal abnormalities.7
The prognosis of RT-DLBCL is generally dismal, but adequate risk stratification may help to guide therapeutic choices. The “Richter syndrome” score from 2006 included performance status, lactate dehydrogenase levels, platelet count, tumor size, and number of previous therapies and was found to predict survival in the chemotherapy era of CLL treatment.8 The previous treatment of CLL is a key prognostic factor with significantly improved survival seen with RT-DLBCL development in previously untreated CLL.9 The Rossi prognostic index from 2011 including TP53 disruption, response to RT-DLBCL treatment, and performance status has also been shown to be highly predictive of survival.10
IgHV analysis of both the underlying CLL and RT-DLBCL is used to determine shared clonality of the disease phases by demonstration of the same heavy-chain variants in both samples, with ∼80% of RT-DLBCL demonstrated as “clonally related” by this method.10 Clonality assessment by comparison of the somatic hypermutation status of the IgHV gene by Sanger or next-generation sequencing as recommended by the European Research Initiative on CLL guidelines11 is considered therapeutically relevant. Several forms of guidance, including these authors’ own recent review, have suggested that allogeneic stem cell transplantation should be considered for all clonally related RT-DLBCL with reservation for use in clonally unrelated RT-DLBCL where initial therapy has failed or where there is coexisting TP53 disruption.1,12,13 In this context, it is noteworthy that assessment of clonal relatedness is sporadic and infrequently performed in routine clinical practice owing to factors such as difficulty in the acquisition of sufficient RT-DLBCL tissue for genomic analysis and a lack of a preceding uncontaminated CLL sample for comparison or routine access to the necessary method of analysis. There remain questions around the day-to-day utility of the abovementioned clonality testing and independent of routinely available key variables, both clinical and molecular. Given these uncertainties, we performed a systematic review focusing on assessment of clonal relationship in RT-DLBCL survival analysis and description of clinical outcomes as delineated by the 2 groups.
Methods
A search using Ovid MEDLINE was conducted between 28 June 2024 and 11 September 2024. The search strategy comprised (1) identification of the relevant disease group using the search terms “Richter” and “transform” and (2) identification of the intervention using the search terms “clonal” and “immunoglobulin.” No publication date restriction was applied but the analysis was restricted to peer-reviewed publications in the English language, and case reports and narrative reviews were excluded from the literature search. The primary outcome of interest was overall survival (OS) of clonally related RT-DLBCL vs clonally unrelated RT-DLBCL compared statistically with a significance test. Secondary outcomes included (1) descriptive statements regarding outcomes of patients with clonally related or clonally unrelated RT-DLBCL, (2) association of clonal relatedness with other biological or clinical factors, and (3) whether these factors were controlled for in the survival analysis.
All studies with outcomes recorded for patients with RT where clonal relatedness was assessed were included. Data were extracted by 2 authors independently (A.B. and C.H.) and compared for concordance. Data were synthesized qualitatively with textual descriptions and quantitatively for the primary outcome. P values of < .05 were considered significant. Critical appraisal of the quality of included studies was conducted using Critical Appraisal Skills Programme tools to identify potential bias, strengths, and limitations. The study was registered on the PROSPERO (International Prospective Register of Systematic Reviews) database (registration number CRD:42024554634).
Results
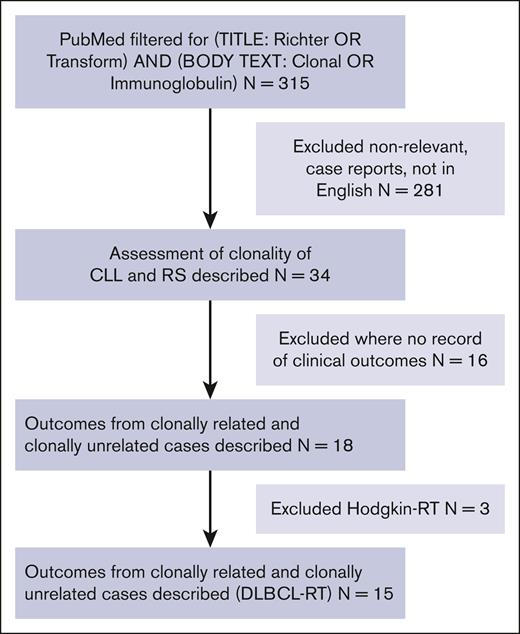
Of 314 manuscripts identified by the search terms (Figure 1), 34 described assessments of clonality of both RT and the underlying CLL to determine clonal relatedness of the 2 disease states. Eighteen of these papers described clinical outcomes associated with clonally related vs clonally unrelated RT. Studies solely examining RT-classical Hodgkin lymphoma were next excluded leaving 15 RT-DLBCL papers for final analysis. Results from these 15 papers are presented in Table 1; where studies solely described the clinical outcomes of patients with no quantitative outcomes, the outcome assessment field has been left blank. Eleven studies assessed outcomes in the first-line RT-DLBCL treatment setting, 2 studies assessed patients with relapsed/refractory (R/R) RT-DLBCL, and 2 studies contained a mixture of first-line and R/R RT-DLBCL. Across the 15 studies that included studies solely reviewing clonality assessment and observational or interventional studies where only a proportion of patients had clonality assessment performed, a total of 747 patients were included, of whom 45% had clonality assessment performed.
Flowchart of study selection from literature review. RS, Richter syndrome.
Comparison of published studies in which clonal relatedness of the underlying CLL and subsequent RT-DLBCL has been examined and where there has been reporting in the study of clinical outcomes as per clonal relatedness
| Author name, year . | Type of study . | Therapy delivered . | Total number of patients in the study . | Number of patients where clonality results available (% clonally related) . | Method of clonality assessment . | Type of outcome assessment . | Outcome assessment . | CASP score—risk of bias . |
|---|---|---|---|---|---|---|---|---|
| Nakamura et al,14 2000 | Observational study at time of diagnosis | Not specified | 3 | 2 (0.0%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Mao et al,15 2007 | Observational study at time of diagnosis | Not specified | 34 | 23 (78.3%) | Immunoglobulin gene rearrangement | Outcome measure not specified but outcomes examined by Cox’s proportional hazards regression model | No survival difference between groups demonstrated | Moderate |
| Rossi et al,10 2011 | Observational study | (R)CHOP, 65% Fludarabine-based, 8.5% Other, 26.5% | 86 | 63 (79.4%) | Immunoglobulin gene rearrangement | Median OS | Significant improvement in clonally unrelated vs clonally related | Low |
| Eyre et al,16 2016 | Interventional study in first-line patients | CHOP + ofatumumab | 37 | 17 (94.1%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Innocenti et al,17 2018 | Observational study at time of diagnosis in patients with ibrutinib-treated CLL | RCHOP | 2 | 2 (100%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Rogers et al,18 2019 | Observational study in R/R patients | PD-1 inhibitors | 10 | 2 (100%) | Not described | Descriptive only—no statistical analysis | N/A | High |
| Abrisqueta et al,19 2020 | Observational study at time of diagnosis | (R)CHOP, 60% Other, 18% None, 22% | 112 | 35 (85.7%) | Immunoglobulin gene rearrangement | Median OS | Significant improvement in clonally unrelated vs clonally related | Moderate |
| Wang et al,9 2020 | Observational study at time of diagnosis | RCHOP, 65% Targeted therapies, 11% Other, 19% None, 5% | 204 | 21 (57.1%) | Immunoglobulin gene rearrangement | Median OS | No significant difference in median OS demonstrated between groups | Moderate |
| Kittai et al,20 2020 | Observational study in R/R patients | Axicabtagene ciloleucel | 9 | 5 (100%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Hess et al,21 2022 | Observational study at time of diagnosis | RCHOP-like | 24 | 7 (85.7%) | Chromosomal microarray analysis | Descriptive only—no statistical analysis | N/A | High |
| Gángó et al,22 2022 | Observational study at time of diagnosis in ibrutinib or patients with venetoclax-treated CLL | Not specified | 6 | 4 (75%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Broséus et al,23 2023 | Observational study at time of diagnosis | RCHOP-like, 87% Platinum based, 7% | 58 | 58 (75.9%) | Genome-wide methylation and whole-transcriptome profiling | Median OS | Significant improvement in clonally unrelated vs clonally related (assessed in 37.9% of entire cohort) | Low |
| Parry et al,7 2023 | Observational study of patients at time of diagnosis and R/R patients | R chemotherapy, 77% Allogeneic, HSCT 8% Autologous HSCT, 7% CAR-T, 3% | 52 | 52 (86.5%) | Whole exome and whole genome sequencing | Median OS | Significant improvement in clonally unrelated vs clonally related | Moderate |
| Wierda et al,24 2024 | Interventional study in first line and R/R patients | Pirtobrutinib | 82 | 21 (85.7%) | Immunoglobulin gene rearrangement | ORR in patients treated with pirtobrutinib—no statistical analysis | Similar between groups (66.67% in clonally unrelated, 61.1% in clonally related) | Moderate |
| Tedeschi et al,25 2024 | Interventional study of patients treated in the first line | Atezolizumab, venetoclax, and obinutuzumab | 28 | 24 (83.3%) | Immunoglobulin gene rearrangement | ORR | No significant difference between groups demonstrated | Moderate |
| Author name, year . | Type of study . | Therapy delivered . | Total number of patients in the study . | Number of patients where clonality results available (% clonally related) . | Method of clonality assessment . | Type of outcome assessment . | Outcome assessment . | CASP score—risk of bias . |
|---|---|---|---|---|---|---|---|---|
| Nakamura et al,14 2000 | Observational study at time of diagnosis | Not specified | 3 | 2 (0.0%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Mao et al,15 2007 | Observational study at time of diagnosis | Not specified | 34 | 23 (78.3%) | Immunoglobulin gene rearrangement | Outcome measure not specified but outcomes examined by Cox’s proportional hazards regression model | No survival difference between groups demonstrated | Moderate |
| Rossi et al,10 2011 | Observational study | (R)CHOP, 65% Fludarabine-based, 8.5% Other, 26.5% | 86 | 63 (79.4%) | Immunoglobulin gene rearrangement | Median OS | Significant improvement in clonally unrelated vs clonally related | Low |
| Eyre et al,16 2016 | Interventional study in first-line patients | CHOP + ofatumumab | 37 | 17 (94.1%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Innocenti et al,17 2018 | Observational study at time of diagnosis in patients with ibrutinib-treated CLL | RCHOP | 2 | 2 (100%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Rogers et al,18 2019 | Observational study in R/R patients | PD-1 inhibitors | 10 | 2 (100%) | Not described | Descriptive only—no statistical analysis | N/A | High |
| Abrisqueta et al,19 2020 | Observational study at time of diagnosis | (R)CHOP, 60% Other, 18% None, 22% | 112 | 35 (85.7%) | Immunoglobulin gene rearrangement | Median OS | Significant improvement in clonally unrelated vs clonally related | Moderate |
| Wang et al,9 2020 | Observational study at time of diagnosis | RCHOP, 65% Targeted therapies, 11% Other, 19% None, 5% | 204 | 21 (57.1%) | Immunoglobulin gene rearrangement | Median OS | No significant difference in median OS demonstrated between groups | Moderate |
| Kittai et al,20 2020 | Observational study in R/R patients | Axicabtagene ciloleucel | 9 | 5 (100%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Hess et al,21 2022 | Observational study at time of diagnosis | RCHOP-like | 24 | 7 (85.7%) | Chromosomal microarray analysis | Descriptive only—no statistical analysis | N/A | High |
| Gángó et al,22 2022 | Observational study at time of diagnosis in ibrutinib or patients with venetoclax-treated CLL | Not specified | 6 | 4 (75%) | Immunoglobulin gene rearrangement | Descriptive only—no statistical analysis | N/A | High |
| Broséus et al,23 2023 | Observational study at time of diagnosis | RCHOP-like, 87% Platinum based, 7% | 58 | 58 (75.9%) | Genome-wide methylation and whole-transcriptome profiling | Median OS | Significant improvement in clonally unrelated vs clonally related (assessed in 37.9% of entire cohort) | Low |
| Parry et al,7 2023 | Observational study of patients at time of diagnosis and R/R patients | R chemotherapy, 77% Allogeneic, HSCT 8% Autologous HSCT, 7% CAR-T, 3% | 52 | 52 (86.5%) | Whole exome and whole genome sequencing | Median OS | Significant improvement in clonally unrelated vs clonally related | Moderate |
| Wierda et al,24 2024 | Interventional study in first line and R/R patients | Pirtobrutinib | 82 | 21 (85.7%) | Immunoglobulin gene rearrangement | ORR in patients treated with pirtobrutinib—no statistical analysis | Similar between groups (66.67% in clonally unrelated, 61.1% in clonally related) | Moderate |
| Tedeschi et al,25 2024 | Interventional study of patients treated in the first line | Atezolizumab, venetoclax, and obinutuzumab | 28 | 24 (83.3%) | Immunoglobulin gene rearrangement | ORR | No significant difference between groups demonstrated | Moderate |
CAR-T, chimeric antigen receptor therapy; CASP, Critical Appraisal Skills Programme; HSCT, hematopoietic stem cell transplant; N/A, not available; ORR, overall response rate; PD-1, programmed death-1; RCHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone; (R)CHOP: (rituximab), cyclophosphamide, doxorubicin, vincristine, and prednisolone.
Laboratory techniques for assessment of clonal relatedness of CLL and RT-DLBCL
The most common laboratory method of assessment of clonal relatedness involved polymerase chain reaction of the immunoglobulin gene sequence of both the underlying CLL and corresponding RT-DLBCL to assess whether both clones have the same IgH variable diversity joining rearrangement. Three studies in our review used different methods. Hess et al21 confirmed clonality of paired CLL and RT-DLBCL samples by chromosomal microarray analysis, which demonstrated fully or partially retained chromosomal aberrations with shared breakpoints and reported outcome data descriptively, in a study too small from which to draw definitive conclusions. Broséus et al23 assessed clonal relationship in 58 patients by genome-wide methylation and whole-transcriptome profiling of the RT-DLBCL with delineation of RT-DLBCL into that which genetically clustered with either CLL or de novo DLBCL. They partially validated their method against traditional IgHV analysis of the RT-DLBCL paired with matched CLL samples in a subset of total cases. This may have significant clinical applicability given that current methods of clonality assessment require a paired CLL sample, which may not always be available. Finally, Parry et al7 used whole exome and whole genome sequencing of paired CLL and RT-DLBCL samples to assess the clonal composition and determined clonal relatedness if at least 1 common ancestral clone was shared between the 2. This method was compared with IgHV analysis of paired samples, and these results in comparison with their novel method were found to be “largely in line.”
Effect of clonal relatedness on survival
Six of the 14 included studies compared median OS between clonally related and clonally unrelated RT-DLBCL with assessment of significance by a statistical test (Table 2).7,9,10,15,19,23 In 4 studies, median OS was compared between groups by Kaplan-Meier analysis with a log-rank test for significance; in 1 study,15 survival was compared between groups using a Cox proportional hazards regression model and in another23 the method of statistical comparison was not described but a P value was given.
Comparison of published studies in which median OS in patients with clonally related and clonally unrelated RT-DLBCL has been compared by a statistical test
| Author name, y . | Intervention . | Number of patients (% clonally related) . | Median OS in clonally related patients . | Median OS in clonally unrelated patients . | Significant difference between groups (P value) . | Potential confounding factors in outcome assessment . |
|---|---|---|---|---|---|---|
| Mao et al,15 2007 | Not specified | 23 (78.3%) | Not specified | Not specified | No (not given) | Related cases were thought to have higher rates of immunoblastic morphology and unmutated IgHV (although not statistically assessed). |
| Rossi et al,10 2011 | RCHOP, 65% Fludarabine-based, 8.5% Other, 26.5% | 63 (79.4%) | 14.2 mo | 62.5 mo | Yes (P = .017) | Related RT group enriched for cases with stereotyped B-cell receptor VH CDR3 (P = .009) and TP53 disruption (P = .018). |
| Abrisqueta et al,19 2020 | RCHOP, 60% Other, 18% None, 22% | 35 (85.7%) | 5.4 mo | 74.8 mo | Yes (P = .05) | Not assessed |
| Wang et al,9 2020 | RCHOP, 65% Targeted therapies, 11% Other, 19% None, 5% | 21 (57.1%) | Not specified | Not specified | No (P = .6) | Not assessed |
| Broséus et al,23 2023 | RCHOP-like, 87% Platinum-based, 7% | 58 (75.9%) | 8 mo | 35.5 mo | Yes (P = .018) | Multivariate analysis confirmed the significance of findings when controlled for IPI, TP53 disruption, and “double-hit” RT. Related RT group was enriched for cases with more heavily pretreated CLL (P = .02), IgHV-unmutated CLL (P = 6.3 ×10–9), and MYD88wt (P = 7 ×10–3). |
| Parry et al,7 2023 | R chemotherapy, 77% Allogeneic HSCT, 8% Autologous HSCT, 7% CAR-T, 3% | 52 (86.5%) | 5.8 mo | 56.4 mo | Yes (P = .0094) | Clonally unrelated RT tended to lack TP53 and NOTCH1 disruption and have underlying CLL, which was IgHV mutated (although not statistically assessed). |
| Author name, y . | Intervention . | Number of patients (% clonally related) . | Median OS in clonally related patients . | Median OS in clonally unrelated patients . | Significant difference between groups (P value) . | Potential confounding factors in outcome assessment . |
|---|---|---|---|---|---|---|
| Mao et al,15 2007 | Not specified | 23 (78.3%) | Not specified | Not specified | No (not given) | Related cases were thought to have higher rates of immunoblastic morphology and unmutated IgHV (although not statistically assessed). |
| Rossi et al,10 2011 | RCHOP, 65% Fludarabine-based, 8.5% Other, 26.5% | 63 (79.4%) | 14.2 mo | 62.5 mo | Yes (P = .017) | Related RT group enriched for cases with stereotyped B-cell receptor VH CDR3 (P = .009) and TP53 disruption (P = .018). |
| Abrisqueta et al,19 2020 | RCHOP, 60% Other, 18% None, 22% | 35 (85.7%) | 5.4 mo | 74.8 mo | Yes (P = .05) | Not assessed |
| Wang et al,9 2020 | RCHOP, 65% Targeted therapies, 11% Other, 19% None, 5% | 21 (57.1%) | Not specified | Not specified | No (P = .6) | Not assessed |
| Broséus et al,23 2023 | RCHOP-like, 87% Platinum-based, 7% | 58 (75.9%) | 8 mo | 35.5 mo | Yes (P = .018) | Multivariate analysis confirmed the significance of findings when controlled for IPI, TP53 disruption, and “double-hit” RT. Related RT group was enriched for cases with more heavily pretreated CLL (P = .02), IgHV-unmutated CLL (P = 6.3 ×10–9), and MYD88wt (P = 7 ×10–3). |
| Parry et al,7 2023 | R chemotherapy, 77% Allogeneic HSCT, 8% Autologous HSCT, 7% CAR-T, 3% | 52 (86.5%) | 5.8 mo | 56.4 mo | Yes (P = .0094) | Clonally unrelated RT tended to lack TP53 and NOTCH1 disruption and have underlying CLL, which was IgHV mutated (although not statistically assessed). |
CAR-T, chimeric antigen receptor therapy; HSCT, hematopoietic stem cell transplant; IPI, International Prognostic Index; MYD88wt, MYD88 wild type; R, rituximab; RCHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone.
In 2 studies with sample sizes of n = 21 and n = 23, a significant difference in median OS between clonally related and clonally unrelated RT-DLBCL could not be demonstrated.9,15
Four other studies comprised a total of 181 patients and reported a median OS of between 5.4 to 14.2 months for clonally related and 35.5 to 74.8 months for clonally unrelated RT-DLBCL.7,19,23,26 A significant difference between the groups was demonstrated in all studies, with descriptive P values ranging between .0094 and .05. Patients in all these 4 studies predominantly received chemoimmunotherapy for treatment of RT-DLBCL.
Two studies in patients with R/R RT and first-line RT assessed the likelihood of overall response rate to treatment (pirtobrutinib24 and atezolizumab, venetoclax, and obinutuzumab25) based on clonal relatedness—neither study found that clonally related or clonally unrelated patients had a differential likelihood of achieving a treatment response. No study has assessed progression-free survival differences in clonally related and clonally unrelated groups.
Interaction of other factors with clonal relatedness and outcomes
Of the 4 analyses demonstrating a significant difference in OS between clonally related and clonally unrelated RT-DLBCL, only 2 have attempted to assess whether other clinical or biological factors may be contributory or confound the results in a statistically meaningful way. Rossi et al,10 with the largest cohort of the series (n = 63), compared the groups for numerous clinical factors including baseline patient, CLL, and RT-DLBCL characteristics and RT-DLBCL treatment by Cox regression analysis and could not demonstrate significant baseline differences between the clonally related and clonally unrelated groups. However, when molecular analysis was taken into account, the clonally related RT-DLBCL group was found to be enriched for cases with stereotyped B-cell receptor VH CDR3 (50% of related vs 7.6% of unrelated, P = .009) and TP53 disruption (60% of related vs 23.1% of unrelated, P = .018). Clonality assessment was included in a multivariate analysis, which demonstrated that performance status, response to therapy, and TP53 mutation status were independent factors associated with OS. In the study by Broséus et al23 (n = 31), clonally related disease was associated with a worse OS and this remained the case when the survival analysis was adjusted for the DLBCL International Prognostic Index, TP53 status, and presence of “double-hit” DLBCL by multivariate analysis using Cox proportional hazards model; however, the clonally related cases also had a significantly higher proportion of patients with more heavily pretreated CLL (66% of unrelated had previously untreated CLL vs 23% of related, P = .02), IgHV unmutated underlying CLL (7% of unrelated vs 93% of related, P = 6.3 × 10–9), and MYD88 wild-type disease (31% of unrelated vs 2% of related, P = 7 × 10–3).
In summary, our literature review has demonstrated that although clonal relatedness of the underlying CLL does confer a poorer survival than clonally unrelated DLBCL-RT, this has not been demonstrated in any study to be independent of all other well-described clinical and genomic variables known to influence outcome in patients with RT-DLBCL.
Discussion
We performed a comprehensive systematic review addressing the important clinical question of whether clonal relatedness of RT-DLBCL to the underlying CLL has demonstrably prognostic value, in the context of a current diagnostic landscape where this is recommended in multiple guidelines but rarely routinely performed. We found a low number of studies (n = 15) where clinical outcomes of clonally related and clonally unrelated RT-DLBCL were contrasted and a lower number still (n = 6) where survival outcomes were compared for statistically significant differences. Four studies comprising 208 patients in total demonstrated statistically significant inferior OS in patients with clonally related CLL, supporting the hypothesis that RT-DLBCL evolving from a preexisting CLL clone is biologically more aggressive and more likely to be treatment resistant than RT-DLBCL, which arises independently of underlying CLL. In 2 studies comprising a total of 44 patients, no significant difference in OS could be demonstrated, although numbers in these studies were small. Among the 4 studies demonstrating a survival difference between groups, 2 used clonality assessment by routinely available IgHV sequencing; however, 2 of these studies used novel unvalidated methods of clonality assessment including genome-wide methylation profiling, whole-transcriptome analysis, and whole exome and genome sequencing, which are clearly some way from adoption for routine use.
We then went on to examine whether clonal relatedness could be deemed a prognostic factor sufficiently strong as to be independent of other commonly available and validated clinical and biological variables, which also denote poor prognosis. Among the 2 eligible studies, the clonally related groups were found to be enriched for cases that expressed the stereotyped B-cell receptor VH CDR3 or were TP53 disrupted, MYD88 wild type, IgHV unmutated, or more heavily pretreated. This suggests that clonal relatedness of RT-DLBCL occurs in combination with other markers for biologically aggressive disease, suggesting that there is a unique biological difference between clonally related and clonally unrelated RT-DLBCL, and underscores that several of these factors should be considered when attempting to predict prognosis in RT-DLBCL. In addition, biological characterization of clonally related and clonally unrelated RT-DLBCL in comparison with de novo DLBCL by genome-wide methylation and whole-transcriptome profiling23 and whole exome/genome sequencing7 demonstrated homogeneity between the biology of clonally unrelated RT-DLBCL and de novo DLBCL, suggesting a distinct origin of clonally unrelated RT-DLBCL that behaves more similarly to its de novo counterpart. To date, clonality assessment alone has not demonstrated strong and complete independence from these other factors in large data sets, particularly in the nonchemotherapy era.
Heterogeneity between treatment of patients included in the studies was seen. Studies describing a significantly poorer survival with clonally related RT-DLBCL mostly included patients treated with chemoimmunotherapy. Interestingly, the patient cohort of 1 study that failed to demonstrate a significant difference between groups comprised 11% treated with targeted therapies9; similarly an interventional study that examined differential response between patients with clonally related and clonally unrelated RT-DLBCL to targeted therapy (atezolizumab, venetoclax, and obinutuzumab) demonstrated no significant difference between groups. This may suggest that clonal relatedness is less likely to be prognostic in patients treated with newer therapies in comparison with traditional chemoimmunotherapy where outcomes in this patient cohort are known to be poor—this possibility requires further investigation in subsequent novel nonchemotherapy trials.
We also interrogated the quality of the studies as part of our review. The small sample size of available studies, with the largest and most recognized case series containing just 63 patients and only 13 clonally unrelated patients, is in keeping with the rarity of RT-DLBCL and the difficulty in applying these laboratory techniques in this patient cohort. Most studies had moderate or high risk of bias as assessed by the Critical Appraisal Skills Programme score, indicating the retrospective analysis involved in most studies and underscoring the need for incorporation of these analyses into prospective clinical trials.
Although not meeting the inclusion criteria for our systematic review, data were presented at the 2024 European Hematology Association from the European Research Initiative on CLL consortium regarding 93 patients with assessment of clonal relatedness (both RT-DLBCL and RT-classical Hodgkin lymphoma).27 The majority (76.3%) were clonally related and these had a poorer outcome than clonally unrelated RT with a median OS of 11.4 months (95% confidence interval, 7.8-19.6) compared with 75.1 months (95% confidence interval, 10.2 to not estimable), respectively (P = .011). Data were not available regarding other prognostic features of the groups.
Even when performed routinely, paired IgHV analysis is not straightforward. Of studies included where clonal relatedness was not the sole factor assessed, paired IgHV analysis by assessment of somatic hypermutation status failed in a significant proportion of patients (11.5%-54.1% across studies), highlighting that information regarding clonal relatedness will be missing in some RT-DLBCL even when this is routinely performed. For paired IgHV analysis of CLL and RT-DLBCL samples to be applicable in practice, there must ideally be a historical record of CLL IgHV mutation type or alternatively a synchronous comparator sample of CLL available; equally, care should be taken to ensure that the RT-DLBCL sample is not contaminated with the CLL clone. Biobank repositories of CLL samples may be helpful for analysis at the time of RT-DLBCL development in the patients who will develop this rare complication but will not be of use for the 10% of patients where RT-DLBCL and CLL present contemporaneously.1 Should this assessment be used to routinely influence patient care, contingency for test failure will have to be incorporated into treatment algorithms. Equally perhaps a more nuanced view of patient prognosis can be undertaken using incorporation of clonal relatedness with other key parameters, eg, TP53 status or previous lines of treatment of CLL.
There are significant challenges with the definition of clonal relatedness as an independent prognostic marker in RT-DLBCL. Assessing for true independent predictive value of this prognostic marker will require much larger cohort studies but the authors recognize these studies are challenging to coordinate. Three of the studies included in our review were interventional clinical trials that included clonality assessment as standard, and we fully support the need for this assessment to be routinely embedded in future prospective interventional trials if it is to shape therapeutic decisions in the future. Newer techniques as described in 2 of the papers may eliminate some of the practical issues surrounding clonality assessment but may be expensive and labor intensive and thus need validation as an independent prognostic factor before implementation in routine clinical care. It is the author’s opinion that further data are required before clonal relatedness of RT-DLBCL is routinely used as a specific diagnostic criterion or indeed as an inclusion criterion within an RT-DLBCL clinical trial.
Authorship
Contribution: A.B. performed a literature search, extracted data, and wrote the manuscript; C.H. performed a literature search and extracted data, which was cross-checked with A.B.’s version for discrepancies and made the visual abstract; and T.A.E. conceptualized the research idea, cross-checked the data extraction, and reviewed and edited the manuscript.
Conflict-of-interest disclosure: A.B. reports receiving travel support to scientific conferences from Ionis Pharmaceuticals, AbbVie, Roche, Karyopharm Therapeutics, and Takeda. T.A.E. reports receiving honorarium and advisory board honorarium from and being in the trial steering committee of Roche; receiving honorarium, research support, and travel support to scientific conferences from Gilead; receiving advisory board honorarium from Kite; receiving travel support to scientific conferences from Takeda; receiving honorarium from Janssen; receiving honorarium and travel support to scientific conferences from AbbVie; receiving honorarium and research funding from and being in the trial steering committee of AstraZeneca; receiving advisory board honorarium from and being in the trial steering committee of Loxo Oncology; receiving advisory board honorarium and research funding from BeiGene; receiving advisory board honorarium and speaker honorarium from Incyte; receiving advisory board honorarium from Autolus; receiving advisory board honorarium from Galapagos; receiving honorarium and advisory board honorarium and being in the trial steering committee of Bristol Myers Squibb; and receiving speaker honoraria from Medscape, PeerView, Clinical Care Options, and The Limbic. C.H. declares no competing financial interests.
Correspondence: Aisling Barrett, Cancer and Haematology Centre, Oxford University Hospitals NHS Foundation Trust, Oxford OX37LE, United Kingdom; email: aisling.barrett@ouh.nhs.uk.
References
Author notes
All patient data contained in this article have been previously published in peer-reviewed journals as contained in the references. Data are available upon reasonable request from the corresponding author, Aisling Barrett (aisling.barrett@ouh.nhs.uk).