Different forms of congenital neutropenia exhibit disruptions in granulocyte development at distinct stages.
There is a strong correlation between the stage and severity of granulocyte development disruption and the efficacy of G-CSF therapy.
Visual Abstract
Congenital neutropenia (CN) is a genetic disorder characterized by persistent or intermittent low peripheral neutrophil counts, thus increasing susceptibility to bacterial and fungal infections. Various forms of CN, caused by distinct genetic mutations, exhibit differential responses to granulocyte colony–stimulating factor (G-CSF) therapy, with the underlying mechanisms not fully understood. This study presents an in-depth comparative analysis of clinical and immunological features in 5 CN patient groups (severe congenital neutropenia [SCN]1, SCN3, cyclic neutropenia [CyN], warts, hypogammaglobulinaemia, infections and myelokathexis [WHIM], and Shwachman-Bodian-Diamond Syndrome [SBDS]) associated with mutations in ELANE, HAX1, CXCR4, and SBDS genes. Our analysis led to the identification of 11 novel mutations in ELANE and 1 each in HAX1, CXCR4, and G6PC3 genes. Investigating bone marrow (BM) granulopoiesis and blood absolute neutrophil count after G-CSF treatment, we found that SCN1 and SCN3 presented with severe early-stage disruption between the promyelocyte and myelocyte, leading to a poor response to G-CSF. In contrast, CyN, affected at the late polymorphonuclear stage of neutrophil development, showed a strong G-CSF response. WHIM, displaying normal neutrophil development, responded robustly to G-CSF, whereas SBDS, with moderate disruption from the early myeloblast stage, exhibited a moderate response. Notably, SCN1 uniquely impeded neutrophil development, whereas SCN3, CyN, WHIM, and SBDS also affected eosinophils and basophils. In addition, SCN1, SCN3, and CyN presented with elevated serum immunoglobulins, increased BM plasma cells, and higher A Proliferation-Inducing Ligand levels. Our study reveals a strong correlation between the stage and severity of granulocyte development disruption and the efficacy of G-CSF therapy.
Introduction
Congenital neutropenia (CN) comprises a range of disorders that are either isolated or part of a complex genetic disease, characterized by a persistent or intermittent, low peripheral neutrophil count (severe <0.5 × 109/L and mild 0.5 - 1.5 × 109/L ).1 The inherited forms of CN often presents with more severe clinical manifestations than those caused by chemotherapy, viral infections, or drug reactions.2 Life-threatening bacterial infections mostly occur in the first year of life for patients with severe congenital neutropenia (SCN) who have not received granulocyte colony–stimulating factor (G-CSF) treatment.3
Granulocyte development proceeds in a stepwise fashion in the bone marrow (BM) and is regulated by various growth factors and cytokines.4 Granulocyte-monocyte progenitor cells, the direct precursors to granulocytes and monocytes, first differentiate into myeloblasts (immature granulocytes) and then into promyelocytes.5 The promyelocytes further differentiate into myelocytes that begin to exhibit distinct characteristics of specific granulocyte lineages, including neutrophils, eosinophils, or basophils.6 The myelocytes mature into metamyelocytes and subsequently into fully formed granulocytes, including band cells and polymorphonuclear leukocytes (PMNs). These cells then leave the BM and migrate to the periphery.7 Patients with SCN exhibit a maturation arrest at the stages of promyelocytes and myelocytes during granulocyte development in the BM.8 However, it is incompletely understood why granulopoiesis is disrupted at specific stage in various types of CN caused by different genetic mutations.
For CN cases with known genetic defects, mutations in the ELANE gene are most prevalent, accounting for 55.6% cases.9 The ELANE (or ELA2) gene encodes the neutrophil elastase protein. A diagnosis of ELANE-related neutropenia is made when a heterozygous pathogenic variant is found in a proband with suggestive clinical findings. Clinical manifestations of ELANE-related neutropenia, which include SCN or cyclic neutropenia (CyN), consist of recurrent fever, skin and oropharyngeal inflammation, and cervical lymphadenitis.10 CN can also arise because of mutations in other genes, including HAX1, CXCR4, and Shwachman-Bodian-Diamond Syndrome (SBDS). These patients require regular G-CSF treatment to prevent frequent infections, but with a risk of ∼15% to 25% of developing myelodysplasia (MDS) or acute myelogenous leukemia.10 Furthermore, patients with CN exhibit heterogeneous responses to G-CSF therapy, but the underlying mechanisms are not well-understood.1
Because ELANE is the most prevalent pathogenic gene in CN, most studies have focused primarily on ELANE-related neutropenia, with few distinguishing it from CN caused by mutations in other genes. To understand the pathogenesis of CN resulting from different genetic defects, we conducted a retrospective analysis of patients with CN at our center and compared the clinical, immunological, and BM characteristics of following 5 CN groups: SCN1 and CyN (both caused by mutations in ELANE), SCN3 (associated with HAX1 mutations), warts, hypogammaglobulinaemia, infections and myelokathexis (WHIM, linked to CXCR4 mutations), and SBDS, related to mutations in the SBDS gene. During this analysis, we discovered 11 novel mutations in ELANE and 1 mutation each in the HAX1, CXCR4, and G6PC3 genes. Notably, we observed distinctive differences in the granulocyte development, G-CSF responsiveness, and antibody production among these 5 groups. Specifically, we identified a strong correlation between the stage and degree of granulocyte development disruption and the efficacy of G-CSF therapy.
Methods
Patients and G-CSF therapy
In this retrospective study conducted at the Department of Clinical Immunology, Children’s Hospital of Fudan University, a total of 349 patients diagnosed with chronic neutropenia were recruited over a period of 8 years (2014-2021). Chronic neutropenia is defined as a decrease in the absolute number of neutrophils in peripheral blood for >3 months. Of these, 56 cases had a diagnosed gene defect. These patients were categorized into 5 groups based on gene screening results and clinical manifestations: SCN1, SCN3, CyN, WHIM, and SBDS. The cases with the causative genes WAS, G6PC3, and SPR54 were not included in the comparative analysis because there was only 1 case each. Typically, patients with SCN were found to have an absolute neutrophil count (ANC) <0.5 × 109 /L for >6 months within the first year of life, accompanied by abnormal granulopoiesis in the BM and frequent monthly infections.
For the diagnosis of CN, routine laboratory examinations were conducted to rule out other potential causes, such as active infections, autoimmune conditions, metabolic disorders, and marrow failure. Complete blood counts were obtained 3 times per week for 6 weeks for CN diagnosis, and the mean neutrophil count was calculated based on ∼10 blood tests, encompassing a comprehensive range of the clinical condition, including any periods of fever or suspected infections. To exclude the influence of G-CSF, the blood routine results of patients in rest state were determined as the mean values of the indicators obtained during their regular follow-up blood routine either before or at least 10 days after the administration of G-CSF. BM aspirations were conducted upon the recommendation of their physicians after diagnosis and before G-CSF treatment. The timing of these aspirations was determined based on the availability and consent of the patients’ parents, thus resulting in random timing relative to the neutrophil cycle. At the time of BM aspiration, the peripheral ANCs in these patients in CyN group were all <1.5 × 109 cells per L threshold for neutropenia diagnosis. The ANCs at the closest time point of the BM sample collection, along with the fluctuation ranges for the 4 patients in CyN group, were as follow: P5 had an ANC of 1.16 × 109 cells per L with a fluctuation range of 0.23 × 109 to 1.84 × 109 cells per L; P11 had an ANC of 0.57 × 109 cells per L with a range of 0.06 × 109 to 2.12 × 109 cells per L; P16 had an ANC of 0.67 × 109 cells per L with a range of 0.10 × 109 to 2.00 × 109 cells per L; P21 had an ANC of 0.46 × 109 cells per L with a range of 0.26 × 109 to 3.73 × 109 cells per L. For each patient, BM aspiration was performed once, and typically 3 BM smears was prepared. A pathologist then counted 200 cells on 1 well-made smear to ascertain the percentages of various cell types. To ensure accuracy, the results were independently verified by another experienced pathologist.
Recombinant human G-CSF (rhG-CSF) was administered at 5 to 20 μg/kg body weight according to patient willingness and risk of infection. Initially, we administered G-CSF at a dose of 5 μg/kg body weight per day for ∼1 week. Subsequently, we increased the doses, typically to 7.5 to 10 μg, based on the initial response observed in patients. In a few cases in which there was no significant increase in neutrophil counts after these doses, we further increased the G-CSF dose, with a maximum dose of 20 μg. The ANC was analyzed before and after G-CSF administration. To address G-CSF responsiveness, we focused on the peak ANC (pANC) achieved within 10 days after G-CSF treatment as our primary criterion.
This study was carried out in accordance with the recommendations of the Ethics Committee of the Children’s Hospital of Fudan University (Shanghai, China) (approval number, 2019-048). Written informed consent was obtained from the parent or guardian, and the child’s assent was secured before any study-related procedures.
Data collection
Clinical and laboratory information was collected via electronic chart review, which included (1) general data, such as gender, age, place of origin, and family history; (2) personal history that focused on the patient’s infection status, including the frequency, type, location, severity, and infection cycle patterns; (3) physical examinations with specific attention to characteristics associated with CN, including skeletal abnormalities, malformation characteristics, albinism, psychomotor development, warts, cardiac function, hepatosplenic lymph node size, and neurological symptoms; (4) laboratory tests encompassed common laboratory screening items, such as blood smear analysis, C-reactive protein levels, serum electrolytes, liver and kidney function, serological and/or DNA or RNA analysis of viral pathogens (cytomegalovirus, Epstein-bar virus, etc), fasting blood glucose, autoantibodies, and BM cytomorphological examination; (5) immunological examinations included lymphocyte subgroup analysis, serum Ig levels, neutrophil respiratory burst function tests, immunoglobulin G (IgG) subclasses, and assessment of specific antibodies; and (6) additional examinations were conducted as needed, including (i) metabolism-related tests, such as urine organic acids, and urine and serum amino acids, to rule out metabolic diseases associated with neutropenia, including glycogen storage disease Ib, organic acidosis, tyrosinemia, Barth syndrome, and Gaucher disease and (ii) microbiological tests based on the clinical sympotoms of infection, such as urine culture, throat wipe culture, blood culture, and/or rapid molecular PCR for bacterial and fungal DNA detection.
Genomic DNA sequencing
Genomic DNA was extracted and sequenced as described.11,12 The concentration and quantity of the DNA samples were measured using a NanoDrop ultraviolet spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Next-generation sequencing was carried out using an immunodeficiency gene panel, which included genes associated with CN, such as AK2, AP3B1, CD40LG, CLPB, CSF3R, CXCR2, CXCR4, DNAJC21, DNM2, DOCK2, EFL1, EIF2AK3, ELANE, G6PC3, GATA1, GATA2, GFI1, GINS1, HAX1, IRAK4, JAGN1, KAT6A, KRAS, LAMTOR2, LYST, MYD88, PGM3, PSTPIP1, RAB27A, RAC2, SBDS, SEC61A1, SLC37A4, SMARCD2, SRP54, STK4, TAZ, TCIRG1, TCN2, TLR8, USB1, VPS13B, VPS45, WAS, WDR1, and WIPF1. Alternatively, whole-exome sequencing was performed. The genomic DNA fragments were enriched by adapter ligations and sequenced on an Illumina HiSeq 2000 platform (Illumina, San Diego, CA). The sequence data were annotated using ANNOVAR and variant effect predictor software, and variant pathogenicity was predicted with SIFT, PolyPhen-2, and MutationTaster tools. For each family, we used either panel sequencing or whole-exome sequencing to identify genetic mutations, and the same method was used for both the patient and their parents. Finally, Sanger sequencing was used to confirm pathogenic mutations.
Statistical analysis
Data were presented using standard parameters, such as the mean, median, interval range, absolute number, standard deviation, and percentile. Categorical variables were shown as n (%), normally distributed continuous variables as mean ± standard deviation, and nonnormally distributed continuous variables as median (interquartile range). For multigroup comparisons, the 1-way analysis of variance test was applied for normally distributed continuous variables, and the Kruskal-Wallis test was used for nonnormally distributed continuous variables in CyN, SCN1, SCN3, WHIM, and SBDS. The 2-way analysis of variance test was used when comparing 2 categorical variables. Data analysis was performed using SPSS 16.0 software or GraphPad Prism 8 software. Statistical significance was evaluated based on the P value (∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001). Gray areas represent reference ranges. For age-related indicators, these ranges are shown as median age reference ranges. The reference ranges used for complete blood counts, BM morphologic subgroups, immunoglobulin subgroups, and lymphocytes are based on our institution’s established norms. For absolute values of lymphocyte subpopulations, refer to the work by Ding et al.13 In addition, the overall proportions of each BM lineage are based on the data provided by Sovani et al.14
Results
Genetic and clinical features of CN
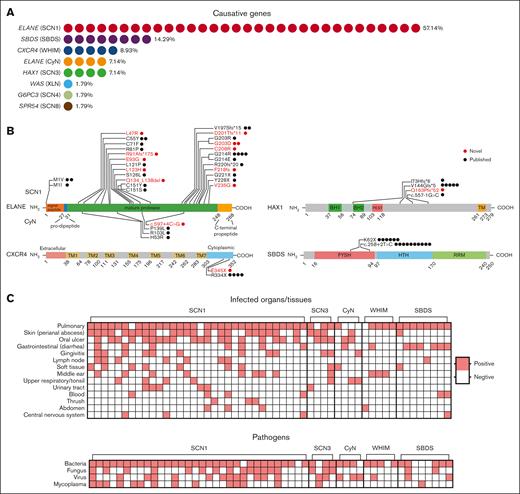
Through a retrospective analysis of patients hospitalized in the Department of Clinical Immunology of the Children’s Hospital of Fudan University from 2014 to 2021, a total of 349 patients with the diagnosis of “chronic neutropenia” were identified. The clinical assessments of our patients were primarily based on their infection histories and routine blood tests. These assessments were conducted both during hospitalization and in outpatient settings. Of the 349 patients, only 56 cases (16%) had a clearly identified causative gene (Figure 1A; Table 1). Patients were categorized into 5 groups based on gene screening results, clinical manifestations, and the number of cases: SCN1 and CyN, both caused by mutations in ELANE; SCN3, associated with HAX1 mutations; WHIM, linked to CXCR4 mutations; and SBDS, related to mutations in the SBDS gene. Cases with the causative genes WAS, G6PC3, and SPR54, which were found in only 1 case each, were excluded from the comparison. The onset age and diagnosis age for each patient group are depicted in supplemental Figure 1A. The majority of these cases were due to ELANE mutations (n = 36, 64.29%), followed by SBDS (n = 814.29%), CXCR4 (n = 5, 8.93%), and HAX1 (n = 4, 7.14%), with only a single case involving mutations in other genes (WAS, G6PC3, and SPR54) (1.79%). We identified 11 novel mutations in ELANE and 1 new mutation each in the HAX1, CXCR4, and G6PC3 genes (Figure 1B, red; Table 2).15-18 De novo mutations emerged as the primary source of ELANE variants, representing 87% of patients with ELANE mutations. This proportion of de novo mutations is consistent with percentages reported in a previous study.19 In addition, ELANE mutations occured either in the first amino acid (M1) or the mature protease region (Figure 1B). Although variants of other genes were mainly inherited within families, only 4 patients (P7, P19-20, and P26) inherited the ELANE variants from their parents (Table 2; supplemental Figure 1B). P7 inherited the p.C55Y variant of ELANE from her father. This variant, which is also present in P6 as a de novo mutation, had been previously reported.15 Siblings P19 and P20 inherited the p.V197Sfs∗15 variant from their father. This mutation was previously reported by Makaryan et al in 2015.16 P26 inherited the p.C208R variant from his father, who had a history suggestive of neutropenia, although exact values were not recorded. The patient’s elder brother also had recurrent respiratory infections in early childhood, but there is no available information about an ELANE mutation.
Genetic and clinical features of CN. (A) A total of 56 cases had a clearly identified causative gene. Among these, 36 had mutations in the ELANE gene, which included SCN1 (n = 32, 57.14%) and CyN (n = 4, 7.14%). The remaining 20 patients had mutations in other genes, which included SBDS (n = 8, 14.29%), CXCR4 (n = 5, 8.93%), HAX1 (n = 4, 7.14%), and WAS, G6PC3, SPR54 with 1 patient each (each accounting for 1.79%). (B) Location of the mutations on the ELANE, HAX1, CXCR4, and SBDS proteins. Each dot represents a case with that particular mutation. Highlighted in red are novel mutations. On the ELANE protein, mutations higher on the diagram are associated with SCN and those lower with CyN. (C) This panel shows the tissues/organs infected or types of infectious agents involved in the medical history of patients in these 5 groups. Red squares indicate a history of infection at the specific site or with the pathogen; White squares denote no reported infection at the site or with the pathogen. BH1 and BH2, Bcl-2 protein homology domains; FYSH, Fungal, Yhr087w, and Shwachman domains; HTH, helix-turn-helix domain; PEST, region rich in proline (P), glutamic acid (E), serine (S), and threonine (T); RRM, RNA recognition motif; TM, transmembrane-like domain.
Genetic and clinical features of CN. (A) A total of 56 cases had a clearly identified causative gene. Among these, 36 had mutations in the ELANE gene, which included SCN1 (n = 32, 57.14%) and CyN (n = 4, 7.14%). The remaining 20 patients had mutations in other genes, which included SBDS (n = 8, 14.29%), CXCR4 (n = 5, 8.93%), HAX1 (n = 4, 7.14%), and WAS, G6PC3, SPR54 with 1 patient each (each accounting for 1.79%). (B) Location of the mutations on the ELANE, HAX1, CXCR4, and SBDS proteins. Each dot represents a case with that particular mutation. Highlighted in red are novel mutations. On the ELANE protein, mutations higher on the diagram are associated with SCN and those lower with CyN. (C) This panel shows the tissues/organs infected or types of infectious agents involved in the medical history of patients in these 5 groups. Red squares indicate a history of infection at the specific site or with the pathogen; White squares denote no reported infection at the site or with the pathogen. BH1 and BH2, Bcl-2 protein homology domains; FYSH, Fungal, Yhr087w, and Shwachman domains; HTH, helix-turn-helix domain; PEST, region rich in proline (P), glutamic acid (E), serine (S), and threonine (T); RRM, RNA recognition motif; TM, transmembrane-like domain.
Clinical features of patients with CN with identified genetic defects diagnosed at Children’s Hospital of Fudan University
| No. . | Gene . | Gender . | Age of onset (mo) . | Age of diagnosis (mo) . | Infected tissues/organs . |
|---|---|---|---|---|---|
| 1 | ELANE | F | 120 | 152 | 1, 7, 11 |
| 2 | ELANE | M | 2 | 3 | 1, 2, 3, 4, 5, 7, 8 |
| 3 | ELANE | M | 3 | 21 | 1, 3, 4, 7, 11 |
| 4 | ELANE | M | 0.4 | 13 | 1, 2, 7 |
| 5 | ELANE(CyN) | F | 9 | 58 | 1, 2, 4, 6 |
| 6 | ELANE | F | 16 | 17 | 1, 11 |
| 7 | ELANE | F | 2 | 33 | 1, 3, 5, 7, 8, 13 |
| 8 | ELANE | F | 0.2 | 17 | 2, 7, 9, 11 |
| 9 | ELANE | M | 5 | 26 | 1, 7 |
| 10 | ELANE | F | 2 | 3 | 1, 2, 3, 6, 7, 10, 11 |
| 11 | ELANE(CyN) | F | 24 | 72 | 2, 8 |
| 12 | ELANE | F | 0.2 | 15 | 1, 2, 3, 5, 7, 8, 10 |
| 13 | ELANE | M | 0.1 | 18 | 1, 2, 10 |
| 14 | ELANE | F | 1 | 162 | 1, 2, 4, 6, 13 |
| 15 | ELANE | M | 2 | 18 | 1, 2, 7 |
| 16 | ELANE(CyN) | F | 24 | 120 | 2, 6, 7 |
| 17 | ELANE | M | 1 | 36 | 1, 3, 7, 11 |
| 18 | ELANE | M | 0.8 | 6 | 1, 4, 5, 7 |
| 19 | ELANE | F | 120 | 152 | 1, 2, 6, 7 |
| 20 | ELANE | M | 2 | 3 | 2, 10 |
| 21 | ELANE(CyN) | F | 3 | 21 | 3, 6 |
| 22 | ELANE | M | 0.4 | 13 | 1, 7, 9, 10, 12 |
| 23 | ELANE | F | 9 | 58 | 1, 2 |
| 24 | ELANE | F | 16 | 17 | 1, 2, 4, 7 |
| 25 | ELANE | F | 2 | 33 | 1, 2, 4, 7, 8, 9, 11, 12 |
| 26 | ELANE | M | 0.2 | 17 | 1 |
| 27 | ELANE | F | 5 | 26 | 1, 2, 3, 5, 8, 11 |
| 28 | ELANE | M | 2 | 3 | 1, 2, 7 |
| 29 | ELANE | M | 24 | 72 | 1, 2, 5, 7 |
| 30 | ELANE | M | 0.2 | 15 | 1, 5, 7, 8, 12 |
| 31 | ELANE | M | 0.1 | 18 | 1, 3, 7 |
| 32 | ELANE | M | 1 | 162 | 1, 2, 3, 5, 7 |
| 33 | ELANE | F | 2 | 18 | 1, 2 |
| 34 | ELANE | F | 24 | 120 | 1 |
| 35 | ELANE | M | 1 | 36 | 1, 2, 7 |
| 36 | ELANE | M | 0.8 | 6 | 1, 8, 9, 13 |
| 37 | HAX1 | M | 3 | 110 | 1, 7, 10 |
| 38 | HAX1 | F | 24 | 69 | 2, 7 |
| 39 | HAX1 | M | 25 | 34 | 1, 2, 3, 7 |
| 40 | HAX1 | M | 1 | 8 | 1, 2, 3, 5, 6, 11 |
| 41 | CXCR4 | M | 24 | 24 | 12 |
| 42 | CXCR4 | M | 2 | 17 | 1, 5, 8 |
| 43 | CXCR4 | M | 0.7 | 13 | 1, 5 |
| 44 | CXCR4 | F | 58 | 62 | 1, 5 |
| 45 | CXCR4 | F | 1 | 24 | 1 |
| 46 | SBDS | F | 120 | 152 | 1, 7, 11 |
| 47 | SBDS | M | 2 | 3 | 1, 8 |
| 48 | SBDS | F | 3 | 21 | 1, 8 |
| 49 | SBDS | F | 0.4 | 13 | 1, 8 |
| 50 | SBDS | F | 9 | 58 | 1, 4 |
| 51 | SBDS | F | 16 | 17 | 1, 8 |
| 52 | SBDS | F | 2 | 33 | 1, 8, 9 |
| 53 | SBDS | M | 0.2 | 17 | 1, 8, 9, 13 |
| 54 | WAS | M | 5 | 26 | 1 |
| 55 | G6PC3 | F | 2 | 3 | 1, 2, 7, 8 |
| 56 | SPR54 | M | 24 | 72 | 1, 2, 4, 11 |
| No. . | Gene . | Gender . | Age of onset (mo) . | Age of diagnosis (mo) . | Infected tissues/organs . |
|---|---|---|---|---|---|
| 1 | ELANE | F | 120 | 152 | 1, 7, 11 |
| 2 | ELANE | M | 2 | 3 | 1, 2, 3, 4, 5, 7, 8 |
| 3 | ELANE | M | 3 | 21 | 1, 3, 4, 7, 11 |
| 4 | ELANE | M | 0.4 | 13 | 1, 2, 7 |
| 5 | ELANE(CyN) | F | 9 | 58 | 1, 2, 4, 6 |
| 6 | ELANE | F | 16 | 17 | 1, 11 |
| 7 | ELANE | F | 2 | 33 | 1, 3, 5, 7, 8, 13 |
| 8 | ELANE | F | 0.2 | 17 | 2, 7, 9, 11 |
| 9 | ELANE | M | 5 | 26 | 1, 7 |
| 10 | ELANE | F | 2 | 3 | 1, 2, 3, 6, 7, 10, 11 |
| 11 | ELANE(CyN) | F | 24 | 72 | 2, 8 |
| 12 | ELANE | F | 0.2 | 15 | 1, 2, 3, 5, 7, 8, 10 |
| 13 | ELANE | M | 0.1 | 18 | 1, 2, 10 |
| 14 | ELANE | F | 1 | 162 | 1, 2, 4, 6, 13 |
| 15 | ELANE | M | 2 | 18 | 1, 2, 7 |
| 16 | ELANE(CyN) | F | 24 | 120 | 2, 6, 7 |
| 17 | ELANE | M | 1 | 36 | 1, 3, 7, 11 |
| 18 | ELANE | M | 0.8 | 6 | 1, 4, 5, 7 |
| 19 | ELANE | F | 120 | 152 | 1, 2, 6, 7 |
| 20 | ELANE | M | 2 | 3 | 2, 10 |
| 21 | ELANE(CyN) | F | 3 | 21 | 3, 6 |
| 22 | ELANE | M | 0.4 | 13 | 1, 7, 9, 10, 12 |
| 23 | ELANE | F | 9 | 58 | 1, 2 |
| 24 | ELANE | F | 16 | 17 | 1, 2, 4, 7 |
| 25 | ELANE | F | 2 | 33 | 1, 2, 4, 7, 8, 9, 11, 12 |
| 26 | ELANE | M | 0.2 | 17 | 1 |
| 27 | ELANE | F | 5 | 26 | 1, 2, 3, 5, 8, 11 |
| 28 | ELANE | M | 2 | 3 | 1, 2, 7 |
| 29 | ELANE | M | 24 | 72 | 1, 2, 5, 7 |
| 30 | ELANE | M | 0.2 | 15 | 1, 5, 7, 8, 12 |
| 31 | ELANE | M | 0.1 | 18 | 1, 3, 7 |
| 32 | ELANE | M | 1 | 162 | 1, 2, 3, 5, 7 |
| 33 | ELANE | F | 2 | 18 | 1, 2 |
| 34 | ELANE | F | 24 | 120 | 1 |
| 35 | ELANE | M | 1 | 36 | 1, 2, 7 |
| 36 | ELANE | M | 0.8 | 6 | 1, 8, 9, 13 |
| 37 | HAX1 | M | 3 | 110 | 1, 7, 10 |
| 38 | HAX1 | F | 24 | 69 | 2, 7 |
| 39 | HAX1 | M | 25 | 34 | 1, 2, 3, 7 |
| 40 | HAX1 | M | 1 | 8 | 1, 2, 3, 5, 6, 11 |
| 41 | CXCR4 | M | 24 | 24 | 12 |
| 42 | CXCR4 | M | 2 | 17 | 1, 5, 8 |
| 43 | CXCR4 | M | 0.7 | 13 | 1, 5 |
| 44 | CXCR4 | F | 58 | 62 | 1, 5 |
| 45 | CXCR4 | F | 1 | 24 | 1 |
| 46 | SBDS | F | 120 | 152 | 1, 7, 11 |
| 47 | SBDS | M | 2 | 3 | 1, 8 |
| 48 | SBDS | F | 3 | 21 | 1, 8 |
| 49 | SBDS | F | 0.4 | 13 | 1, 8 |
| 50 | SBDS | F | 9 | 58 | 1, 4 |
| 51 | SBDS | F | 16 | 17 | 1, 8 |
| 52 | SBDS | F | 2 | 33 | 1, 8, 9 |
| 53 | SBDS | M | 0.2 | 17 | 1, 8, 9, 13 |
| 54 | WAS | M | 5 | 26 | 1 |
| 55 | G6PC3 | F | 2 | 3 | 1, 2, 7, 8 |
| 56 | SPR54 | M | 24 | 72 | 1, 2, 4, 11 |
P19 and P20, as well as P28 and P29, are from 2 separate twin families.
Infected tissues/organs: 1, pulmonary; 2, oral; 3, gums; 4, lymph nodes; 5, middle ear; 6, tonsils; 7, skin; 8, gastrointestinal tract; 9, blood; 10, urinary tract; 11, soft tissues; 12, abdominal cavity; 13, brain/nerves.
F, female. M, male.
Detailed gene variants information in genetically defined CN
| No. . | Gene . | Location . | Nucleotide change . | Protein level . | Inheritance . | Novel mutation∗ . | Clinical significance† . | Reference (PMID‡) . |
|---|---|---|---|---|---|---|---|---|
| 1 | ELANE | Exon1 | c.1A>G | p.M1V | De novo | No | Pathogenic | 24184683 |
| 2 | ELANE | Exon1 | c.1A>G | p.M1V | De novo | No | Pathogenic | 24184683 |
| 3 | ELANE | Exon1 | c.3G>A | p.M1I | De novo | No | Pathogenic | 24184683 |
| 4 | ELANE | Exon2 | c.140T>G | p.L47R | De novo | Yes | Pathogenic | NA§ |
| 5‖ | ELANE | Exon2 | c.158A>G | p.H53R | De novo | No | Pathogenic | NA§ |
| 6 | ELANE | Exon2 | c.164G>A | p.C55Y | NA§ | No | Pathogenic | 16986121 |
| 7 | ELANE | Exon2 | c.164G>A | p.C55Y | Paternal | No | Pathogenic | 16986121 |
| 8 | ELANE | Exon2 | c.212G>T | p.C71F | De novo | No | Pathogenic | 31839986 |
| 9 | ELANE | Exon3 | c.242G>C | p.R81P | De novo | No | Likely Pathogenic | 16986121 |
| 10¶ | ELANE | Exon3 | c.269_277del | p.R91Afs∗175 | De novo | Yes | NA§ | NA§ |
| ELANE | Exon3 | c.278A>G | p.E93G | De novo | Yes | Pathogenic | NA§ | |
| 11‖ | ELANE | Exon3 | c.308G>T | p.R103L | De novo | No | Likely Pathogenic | 17761833 |
| 12 | ELANE | Exon3 | c.362T>C | p.L121P | De novo | No | Likely Pathogenic | NA§ |
| 13 | ELANE | Exon4 | c.368T>A | p.L123H | NA | Yes | Pathogenic | NA§ |
| 14 | ELANE | Exon4 | c.377C>T | p.S126L | De novo | No | Pathogenic | 28492532, 11001877, 14962902, 16079102, 16737875, 18611981, 20582973, 22758217, 23463630, 16551967, 26567890 |
| 15 | ELANE | Exon4 | c.401_415del | p.Q134 _L138del | De novo | Yes | NA§ | NA§ |
| 16‖ | ELANE | Exon4 | c.416C>T | p.P139L | NA | No | Pathogenic | 11001877, 23463630, 21425445, 14962902, 30040071, 16079102, 31321910, 31248972 |
| 17 | ELANE | Exon4 | c.452G>A | p.C151Y | NA§ | No | Pathogenic | 24523240, 25427142, 11675333, 23463630 |
| 18 | ELANE | Exon4 | c.452G>C | p.C151S | De novo | No | Pathogenic | 23463630 |
| 19 | ELANE | Exon4 | c.588delC | p.V197Sfs∗15 | Paternal | No | NA§ | 25427142 |
| 20 | ELANE | Exon4 | c.588delC | p.V197Sfs∗15 | Paternal | No | NA§ | 25427142 |
| 21‖ | ELANE | Exon4 | c.597+4C>G | splicing | NA§ | Yes | NA§ | NA§ |
| 22 | ELANE | Exon5 | c.601del | p.D201Tfs∗11 | De novo | Yes | NA§ | NA§ |
| 23 | ELANE | Exon5 | c.607G>C | p.G203R | NA§ | No | Pathogenic | 23463630 |
| 24 | ELANE | Exon5 | c.608G>A | p.G203D | De novo | Yes | Pathogenic | NA§ |
| 25 | ELANE | Exon5 | c.608G>A | p.G203D | De novo | Yes | Pathogenic | NA§ |
| 26 | ELANE | Exon5 | c.622T>C | p.C208R | paternal | Yes | Pathogenic | NA§ |
| 27 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 28 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 29 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 30 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 31 | ELANE | Exon5 | c.641G>A | p.G214E | De novo | No | Likely Pathogenic | NA§ |
| 32 | ELANE | Exon5 | c.651delC | p.F218fs | De novo | Yes | NA | NA§ |
| 33 | ELANE | Exon5 | c.658delC | p.R220fs∗20 | De novo | No | Pathogenic | 26174650 |
| 34 | ELANE | Exon5 | c.661G>T | p.G221X | De novo | No | Uncertain Significance | 11001877 |
| 35 | ELANE | Exon5 | c.684C>G | p.Y228X | De novo | No | NA§ | 32054657 |
| 36 | ELANE | Exon5 | c.704T>G | p.V235G | De novo | Yes | Pathogenic | NA |
| 37 | HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Maternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 |
| HAX1 | Exon5 | c.557-1G>C | splicing | Paternal | No | Likely Pathogenic | 16199547, 17187068 | |
| 38 | HAX1 | Exon2 | c.216_217insC | p.I73Hfs∗6 | Maternal | No | Pathogenic | 17187068 |
| HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Paternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 | |
| 39 | HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Maternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 |
| HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Paternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 | |
| 40 | HAX1 | Exon3 | c.486_487dup | p.Q163Pfs∗52 | Maternal | Yes | NA§ | NA§ |
| HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Paternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 | |
| 41 | CXCR4 | Exon2 | c.1000C>T | p.R334X | NA | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 42 | CXCR4 | Exon2 | c.1000C>T | p.R334X | NA | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 43 | CXCR4 | Exon2 | c.1000C>T | p.R334X | NA | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 44 | CXCR4 | Exon2 | c.1032dupT | p.E345X | De novo | Yes | NA§ | NA§ |
| 45 | CXCR4 | Exon2 | c.1000C>T | p.R334X | De novo | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 46 | SBDS | Exon2 | c.184A>T | p.K62X | Maternal | No | Pathogenic/likely pathogenic | 33607811 37885353 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 47 | SBDS | Exon2 | c.184A>T | p.K62X | Maternal | No | Pathogenic/likely pathogenic | 33607811 37885353 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 48 | SBDS | Intron2 | c.258+2T>C | splicing | Maternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 49 | SBDS | Intron2 | c.258+2T>C | splicing | Maternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 50 | SBDS | Exon2 | c.183_184TA>CT | p.K62X | Maternal | No | Pathogenic | 12496757, 15769891, 32412173 |
| SBDS | Exon2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 51 | SBDS | Intron2 | c.258+2T>C | splicing | Maternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 |
| SBDS | Exon2 | c.183_184TA>CT | p.K62X | Paternal | No | Pathogenic | 12496757, 15769891, 32412173 | |
| 52 | SBDS | Exon2 | c.184A>T | p.K62X | Maternal | No | Pathogenic/likely pathogenic | 33607811 37885353 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 53 | SBDS | Exon2 | c.183_184TA>CT | p.K62X | Maternal | No | Pathogenic | 12496757, 15769891, 32412173 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 54 | WAS | Exon9 | c.881T>C | p.I294T | Maternal | No | Pathogenic/likely pathogenic | 11242115, 19006568 |
| 55 | G6PC3 | Exon3 | c.394C>T | p.Q132X | Maternal | Yes | NA | NA |
| 56 | SPR54 | Exon5 | c.349_351del | p.T117del | De novo | No | Pathogenic | 29914977, 36159802 |
| No. . | Gene . | Location . | Nucleotide change . | Protein level . | Inheritance . | Novel mutation∗ . | Clinical significance† . | Reference (PMID‡) . |
|---|---|---|---|---|---|---|---|---|
| 1 | ELANE | Exon1 | c.1A>G | p.M1V | De novo | No | Pathogenic | 24184683 |
| 2 | ELANE | Exon1 | c.1A>G | p.M1V | De novo | No | Pathogenic | 24184683 |
| 3 | ELANE | Exon1 | c.3G>A | p.M1I | De novo | No | Pathogenic | 24184683 |
| 4 | ELANE | Exon2 | c.140T>G | p.L47R | De novo | Yes | Pathogenic | NA§ |
| 5‖ | ELANE | Exon2 | c.158A>G | p.H53R | De novo | No | Pathogenic | NA§ |
| 6 | ELANE | Exon2 | c.164G>A | p.C55Y | NA§ | No | Pathogenic | 16986121 |
| 7 | ELANE | Exon2 | c.164G>A | p.C55Y | Paternal | No | Pathogenic | 16986121 |
| 8 | ELANE | Exon2 | c.212G>T | p.C71F | De novo | No | Pathogenic | 31839986 |
| 9 | ELANE | Exon3 | c.242G>C | p.R81P | De novo | No | Likely Pathogenic | 16986121 |
| 10¶ | ELANE | Exon3 | c.269_277del | p.R91Afs∗175 | De novo | Yes | NA§ | NA§ |
| ELANE | Exon3 | c.278A>G | p.E93G | De novo | Yes | Pathogenic | NA§ | |
| 11‖ | ELANE | Exon3 | c.308G>T | p.R103L | De novo | No | Likely Pathogenic | 17761833 |
| 12 | ELANE | Exon3 | c.362T>C | p.L121P | De novo | No | Likely Pathogenic | NA§ |
| 13 | ELANE | Exon4 | c.368T>A | p.L123H | NA | Yes | Pathogenic | NA§ |
| 14 | ELANE | Exon4 | c.377C>T | p.S126L | De novo | No | Pathogenic | 28492532, 11001877, 14962902, 16079102, 16737875, 18611981, 20582973, 22758217, 23463630, 16551967, 26567890 |
| 15 | ELANE | Exon4 | c.401_415del | p.Q134 _L138del | De novo | Yes | NA§ | NA§ |
| 16‖ | ELANE | Exon4 | c.416C>T | p.P139L | NA | No | Pathogenic | 11001877, 23463630, 21425445, 14962902, 30040071, 16079102, 31321910, 31248972 |
| 17 | ELANE | Exon4 | c.452G>A | p.C151Y | NA§ | No | Pathogenic | 24523240, 25427142, 11675333, 23463630 |
| 18 | ELANE | Exon4 | c.452G>C | p.C151S | De novo | No | Pathogenic | 23463630 |
| 19 | ELANE | Exon4 | c.588delC | p.V197Sfs∗15 | Paternal | No | NA§ | 25427142 |
| 20 | ELANE | Exon4 | c.588delC | p.V197Sfs∗15 | Paternal | No | NA§ | 25427142 |
| 21‖ | ELANE | Exon4 | c.597+4C>G | splicing | NA§ | Yes | NA§ | NA§ |
| 22 | ELANE | Exon5 | c.601del | p.D201Tfs∗11 | De novo | Yes | NA§ | NA§ |
| 23 | ELANE | Exon5 | c.607G>C | p.G203R | NA§ | No | Pathogenic | 23463630 |
| 24 | ELANE | Exon5 | c.608G>A | p.G203D | De novo | Yes | Pathogenic | NA§ |
| 25 | ELANE | Exon5 | c.608G>A | p.G203D | De novo | Yes | Pathogenic | NA§ |
| 26 | ELANE | Exon5 | c.622T>C | p.C208R | paternal | Yes | Pathogenic | NA§ |
| 27 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 28 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 29 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 30 | ELANE | Exon5 | c.640G>A | p.G214R | De novo | No | Pathogenic | 11001877, 15657182, 16079102, 28073911, 30386760, 3229910 |
| 31 | ELANE | Exon5 | c.641G>A | p.G214E | De novo | No | Likely Pathogenic | NA§ |
| 32 | ELANE | Exon5 | c.651delC | p.F218fs | De novo | Yes | NA | NA§ |
| 33 | ELANE | Exon5 | c.658delC | p.R220fs∗20 | De novo | No | Pathogenic | 26174650 |
| 34 | ELANE | Exon5 | c.661G>T | p.G221X | De novo | No | Uncertain Significance | 11001877 |
| 35 | ELANE | Exon5 | c.684C>G | p.Y228X | De novo | No | NA§ | 32054657 |
| 36 | ELANE | Exon5 | c.704T>G | p.V235G | De novo | Yes | Pathogenic | NA |
| 37 | HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Maternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 |
| HAX1 | Exon5 | c.557-1G>C | splicing | Paternal | No | Likely Pathogenic | 16199547, 17187068 | |
| 38 | HAX1 | Exon2 | c.216_217insC | p.I73Hfs∗6 | Maternal | No | Pathogenic | 17187068 |
| HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Paternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 | |
| 39 | HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Maternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 |
| HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Paternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 | |
| 40 | HAX1 | Exon3 | c.486_487dup | p.Q163Pfs∗52 | Maternal | Yes | NA§ | NA§ |
| HAX1 | Exon3 | c.430dupG | p.V144Gfs∗5 | Paternal | No | Pathogenic | 28492532, 17187068, 18337561, 20065084, 20220065, 22102707, 24482108 | |
| 41 | CXCR4 | Exon2 | c.1000C>T | p.R334X | NA | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 42 | CXCR4 | Exon2 | c.1000C>T | p.R334X | NA | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 43 | CXCR4 | Exon2 | c.1000C>T | p.R334X | NA | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 44 | CXCR4 | Exon2 | c.1032dupT | p.E345X | De novo | Yes | NA§ | NA§ |
| 45 | CXCR4 | Exon2 | c.1000C>T | p.R334X | De novo | No | Pathogenic | 31313072, 12692554, 25662009, 31493092 |
| 46 | SBDS | Exon2 | c.184A>T | p.K62X | Maternal | No | Pathogenic/likely pathogenic | 33607811 37885353 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 47 | SBDS | Exon2 | c.184A>T | p.K62X | Maternal | No | Pathogenic/likely pathogenic | 33607811 37885353 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 48 | SBDS | Intron2 | c.258+2T>C | splicing | Maternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 49 | SBDS | Intron2 | c.258+2T>C | splicing | Maternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 50 | SBDS | Exon2 | c.183_184TA>CT | p.K62X | Maternal | No | Pathogenic | 12496757, 15769891, 32412173 |
| SBDS | Exon2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 51 | SBDS | Intron2 | c.258+2T>C | splicing | Maternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 |
| SBDS | Exon2 | c.183_184TA>CT | p.K62X | Paternal | No | Pathogenic | 12496757, 15769891, 32412173 | |
| 52 | SBDS | Exon2 | c.184A>T | p.K62X | Maternal | No | Pathogenic/likely pathogenic | 33607811 37885353 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 53 | SBDS | Exon2 | c.183_184TA>CT | p.K62X | Maternal | No | Pathogenic | 12496757, 15769891, 32412173 |
| SBDS | Intron2 | c.258+2T>C | splicing | Paternal | No | Pathogenic | 12496757, 22935661, 21695142, 14749921 | |
| 54 | WAS | Exon9 | c.881T>C | p.I294T | Maternal | No | Pathogenic/likely pathogenic | 11242115, 19006568 |
| 55 | G6PC3 | Exon3 | c.394C>T | p.Q132X | Maternal | Yes | NA | NA |
| 56 | SPR54 | Exon5 | c.349_351del | p.T117del | De novo | No | Pathogenic | 29914977, 36159802 |
Novel variant denotes that the variant was not reported in the NCBI website ClinVar database and GnomAD database. We also referred to ELANE mutations published previously.15-18
Clinical significance represents the assessment of the variant in the ClinVar database or the predicted pathogenicity using polyphen-2.
Reference PMID represents the number of the reported or referenced literature in Pubmed for the mutation.
NA, not available.
Cyclic neutropenia (CyN).
Compound heterozygous mutations. P19 and P20, as well as P28 and P29, are from 2 separate twin families. The father of P19 and P20 has the same compound heterozygous mutations and presented with neutropenia, recurrent respiratory infections, and oral ulcers. P26 inherited the p.C208R variant from his father, whose blood tests were suggestive of neutropenia, although the exact neutrophil count was not recorded.
Recurrent multisite infections by multiple pathogens are the most common clinical manifestations in patients with CN.8,20 In terms of infected organs or tissues, pulmonary infections were present in >80% of cases across all groups except for CyN (Figure 1C, upper panel). Skin infections (mostly perianal abscesses) and oral ulcers occurred more frequently in SCN1, SCN3, and CyN, whereas SBDS cases manifested more gastrointestinal infections (incuding diarrhea) and WHIM cases exhibited more otitis media. Patients who exhibited skin infections or oral ulcers were prescribed antibiotics without hospitalization. With respect to the pathogens involved (Figure 1C, lower panel), bacterial infections were the most dominant pathogens, and fungal infections occurring more frequently in SCN1 and SCN3. The most commonly identified bacteria species were Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli (supplemental Figure 1C). Candida albicans was the most commonly identified fungus, and Mycoplasma pneumoniae was the predominant mycoplasma identified. There was no specific trend observed for viral infections. Among the 56 cases studied, 40 cases received G-CSF treatment. Partly because of the relatively short follow-up period after admission, we were unaware of any cases of leukemia development. However, there was 1 patient who died of severe infections.
Comparative analysis of immune cells and granulocyte subsets in peripheral blood and BM
Firstly, we analyzed the absolute counts of white blood cells (WBCs) along with the 3 major lineages (granulocytes, monocytes, and lymphocytes) in peripheral blood. Given the functional nature of CXCR4 as a chemokine,21,CXCR4 mutation severely affects the release of BM lymphocytes into the periphery, as observed in WHIM syndrome.22,23 Indeed, as depicted in Figure 2A, WHIM displayed a markedly low WBC count because of a significant reduction in monocytes and lymphocytes in addition to low levels of granulocytes. In addition, SCN1 and SBDS showed slightly higher peripheral blood WBC counts than SCN3 and CyN. This could be attributable to an increased count of lymphocytes in SCN1 and SBDS (Figure 2A). Monocytes were increased in SCN1 and SCN3 (Figure 2A), which exceeded the normal reference range. Within the granulocytes, neutrophil counts were higher in CyN and SBDS, whereas eosinophils and basophils were increased in SCN1 (Figure 2B). These observations suggest that SCN1 primarily results in a significant decrease in neutrophils but not in eosinophils and basophils, whereas other genetic variants affect all types of granulocytes. One patient in CyN group (P21, mean ANC = 1.7 × 109 cells per L) and 3 patients in SBDS group (P47, mean ANC = 1.6 × 109 cells per L; P50, mean ANC = 2.05 × 109 cells per L; P52, mean ANC = 1.57 × 109 cells per L) had neutrophil counts slightly >1.5 × 109 cells per L. The ELANE mutation in P21 (ANC range, 0.26-3.73 × 109 cells per L) is novel. However, this mutation, c.597+4G>A, is in close proximity to other mutations (c.597+1G>A, c.597+5G>A) that have been associated with CyN or SCN in the ClinVar database, suggesting a potential pathogenic nature for the P21 mutation. All 3 SBDS mutations have been previously reported as pathogenic. The relevant details can be found in Table 2 (PMID 33607811, 37885353, 12496757, 22935661, 21695142, 14749921, 15769891, and 32412173).
Comparative analysis of BM or peripheral blood immune cell subsets across 5 groups using routine blood tests and cell morphology assays of BM smears. (A) Total number of WBC, granulocytes, monocytes, and lymphocytes in the peripheral blood. (B) Absolute numbers of granulocytes (neutrophils, eosinophils, and basophils) in the peripheral blood. The brown line in neutrophil indicates the cut-off value of 1.5 × 109 cells per L of neutropenia. (C) Proportion of granulocytes, monocytes, and lymphocytes in BM. (D) Proportion of specific granulocyte types (neutrophils, eosinophils, and basophils) in BM. (E-G) Proportions of BM neutrophilic (E-F) and eosinophilic (E,G) lineage cells at different developmental stages for each patient group. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Kruskal-Wallis test was used for comparison among 5 groups. Bars represent means ± standard deviation [SD]. The gray area indicates reference range of each index, as described in the Methods section.
Comparative analysis of BM or peripheral blood immune cell subsets across 5 groups using routine blood tests and cell morphology assays of BM smears. (A) Total number of WBC, granulocytes, monocytes, and lymphocytes in the peripheral blood. (B) Absolute numbers of granulocytes (neutrophils, eosinophils, and basophils) in the peripheral blood. The brown line in neutrophil indicates the cut-off value of 1.5 × 109 cells per L of neutropenia. (C) Proportion of granulocytes, monocytes, and lymphocytes in BM. (D) Proportion of specific granulocyte types (neutrophils, eosinophils, and basophils) in BM. (E-G) Proportions of BM neutrophilic (E-F) and eosinophilic (E,G) lineage cells at different developmental stages for each patient group. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Kruskal-Wallis test was used for comparison among 5 groups. Bars represent means ± standard deviation [SD]. The gray area indicates reference range of each index, as described in the Methods section.
To explore the mechanisms underlying the differential distribution of peripheral immune cell subsets, we next performed a detailed analysis of the development of distinct immune cell lineages in BM. We analyzed the proportions of the 3 lineages (granulocytes, monocytes, and lymphocytes) in the BM (Figure 2C). Both SCN1 and SCN3 showed significantly lower proportions of the granulocyte lineage compared with CyN and WHIM (Figure 2C), predominantly because of a drastic decrease in the neutrophil lineage (Figure 2D). In contrast, CyN and WHIM showed higher proportions of the granulocyte lineages than other subgroups (Figure 2C), largely because of the normal development of neutrophilic granulocytes in the BM (Figure 2D). Within the granulocyte lineage, the proportion of neutrophil lineage was reduced, whereas those of eosinophil and basophil lineages increased in SCN1 (Figure 2D). These observations are in agreement with the findings in the peripheral blood (Figure 2B).
We further examined the specific stages of neutrophil development in the BM (Figure 2E-G). SCN1 and SCN3 exhibited severe dysplasia beginning at the commitment to the neutrophilic lineage (myelocytes) and persisting through to the PMNs (Figure 2F). In contrast, CyN displayed relatively normal development of neutrophilic granulocytes up to the band cell stage, but with a moderate reduction in PMNs (Figure 2F). Neutrophil development in SBDS was intermediate between these 2 patterns. WHIM showed normal neutrophil development from the myelocyte all the way through to the PMNs. These results demonstrate that neutrophilic granulocyte development is variably affected among these groups. Unlike the neutrophilic granulocytes, eosinophilic granulocyte development in SCN1 was almost unaffected, whereas it was severely impaired in WHIM and SBDS, from eosinophilic myelocytes to the eosinophilic PMNs (Figure 2G). Meanwhile, eosinophil development in SCN3 and CyN was normal until the band cell stage but became affected at the PMN stage. Thus, there are entirely opposing effects on the development of neutrophilic vs eosinophilic granulocytes in SCN1, resulting in reduced peripheral neutrophils but normal eosinophils (Figure 2B).
Distinct responsiveness to G-CSF among different types of CN
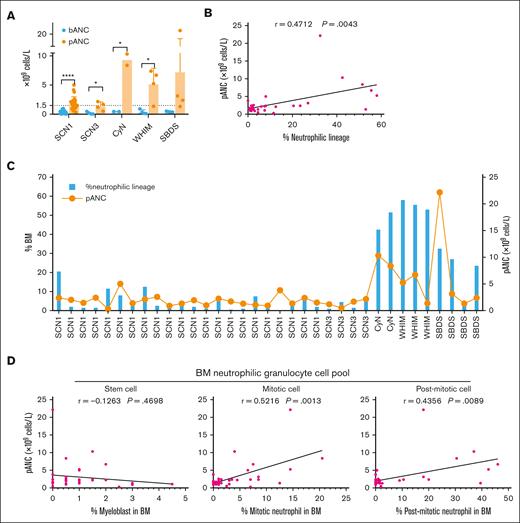
Considering the different impacts of various genetic mutations on the counts of BM and peripheral neutrophils, we further analyzed the effects of G-CSF administration on patients in different groups (Table 3). G-CSF was administered as described in the “Methods.” To address G-CSF responsiveness, we chose to focus on the pANC achieved within 10 days after G-CSF treatment as our primary criterion. It was found that all patients with CN exhibited significantly higher pANC after G-CSF treatment than baseline ANC (bANC) before G-CSF initiation. However, although ANCs were uniformly very low in all patient groups before G-CSF administration, pANC reached different levels in different groups. Notably, pANC was higher in the CyN, WHIM, and SBDS groups, suggesting better responsiveness to G-CSF, whereas only half of the SCN1 and SCN3 were able to elevate their pANC >1.5 × 109 cells per L (Figure 3A; Table 4). This indicates that although G-CSF is effective for all patients with CN, the degree of responsiveness varies among different types of CN.
G-CSF administration and follow-up of patients with CN
| No. . | Mutated gene . | Age at first visit (y) . | Follow-up period (mo) . | Body weight (kg) . | G-CSF . | HSCT∗ . | |||
|---|---|---|---|---|---|---|---|---|---|
| Injection period (d) . | Dosage (μg/kg) . | bANC (×109 cells per L)† . | pANC (×109 cells per L)‡ . | ||||||
| 1 | ELANE | 13.5 | 1.6 | 33.5 | 9 | 7 | 0.53 | 2.37 | No |
| 2 | ELANE | 1.1 | 2.0 | 10.0 | 59 | 10-20 | 0.03 | 0.45 | No |
| 3 | ELANE | 1.8 | 20.8 | 12.5 | 14 | 10 | 0.65 | 2.01 | Yes |
| 4 | ELANE | 1.1 | 3.0 | 10 | — | — | — | — | No |
| 5 | ELANE(CyN) | 4.8 | 1.7 | 15 | — | — | — | — | No |
| 6 | ELANE | 1.4 | 2.3 | 11.5 | — | — | — | — | No |
| 7 | ELANE | 2.7 | 6.5 | 12 | 11 | 10 | 0.32 | 1.45 | Yes |
| 8 | ELANE | 1.4 | 2.4 | 9.5 | 9 | 5-10 | 0.34 | 2.41 | No |
| 9 | ELANE | 1.0 | 80.3 | 11.5 | 5 | 5-7.5 | 0.08 | 0.30 | No |
| 10 | ELANE | 0.2 | 8.1 | 6.6 | 15 | 7.5 | 0.47 | 5.01 | Yes |
| 11 | ELANE(CyN) | 6.7 | 0.5 | 23 | — | — | — | — | No |
| 12 | ELANE | 0.2 | 113.4 | 10 | 7 | 5-20 | 0.24 | 1.37 | No |
| 13 | ELANE | 1.5 | 7.8 | NA | — | — | — | — | No |
| 14 | ELANE | 13.4 | 12.0 | NA | — | — | — | — | Yes |
| 15 | ELANE | 1.5 | 29.9 | 12.5 | 53 | 7.5-15 | 0.91 | 2.11 | Yes |
| 16 | ELANE(CyN) | 10.1 | 2.1 | 26 | 10 | 5 | 0.45 | 10.33 | No |
| 17 | ELANE | 3.1 | 33.1 | 18 | 3 | 5 | 0.40 | 2.57 | Yes |
| 18 | ELANE | 0.5 | 0.5 | 6 | 9 | 10-15 | 0.04 | 0.90 | No |
| 19 | ELANE | 9.1 | 0.6 | 32 | 3 | 5 | 0.68 | 4.17 | No |
| 20 | ELANE | 5.7 | 0.6 | 20 | 11 | 5-10 | 0.26 | 1.32 | No |
| 21 | ELANE(CyN) | 0.4 | 33.5 | 15 | 1 | 5 | 0.46 | 8.38 | No |
| 22 | ELANE | 0.4 | 11.4 | 8 | 9 | 5 | 0.33 | 1.94 | No |
| 23 | ELANE | 2.5 | 0.9 | 11.5 | — | — | — | — | No |
| 24 | ELANE | 1.4 | 0.3 | 10 | — | — | — | — | Yes |
| 25 | ELANE | 1.0 | 60.0 | 9 | 93 | 5-20 | 0.05 | 0.98 | Yes |
| 26 | ELANE | 5.0 | 51.4 | 17 | 3 | 5 | 0.40 | 2.24 | No |
| 27 | ELANE | 2.5 | 19.6 | 12 | 50 | 5-7.5 | 0.18 | 1.71 | Yes |
| 28 | ELANE | 0.5 | 12.3 | 6 | 3 | 5 | 0.96 | 1.30 | No |
| 29 | ELANE | 0.8 | 1.5 | 8 | 4 | 5 | 0.90 | 1.07 | No |
| 30 | ELANE | 1.8 | 38.5 | 10 | 46 | 5-7.5 | 0.55 | 0.92 | Yes |
| 31 | ELANE | 6.5 | 15.4 | 16 | 9 | 5 | 0.42 | 0.55 | Yes |
| 32 | ELANE | 3.0 | 1.6 | 13 | 1 | 5 | 0.20 | — | No |
| 33 | ELANE | 1.1 | 1.4 | 10 | 10 | 5-10 | 0.11 | 3.81 | Yes |
| 34 | ELANE | 13.2 | 0.6 | 12 | 6 | 5-10 | 0.43 | 1.34 | No |
| 35 | ELANE | 6.6 | 3.3 | 22 | 7 | 5-10 | 0.18 | 2.37 | Yes |
| 36 | ELANE | 0.1 | 15.3 | 3 | 7 | 10-15 | 0.08 | 1.50 | No |
| 37 | HAX1 | 1.0 | 13.7 | 10 | 9 | 5-15 | 0.06 | 1.19 | No |
| 38 | HAX1 | 3.7 | 1.2 | 11 | 2 | 5 | 0.03 | 0.51 | No |
| 39 | HAX1 | 10.9 | 56.5 | 31 | 5 | 5 | 0.22 | 1.68 | No |
| 40 | HAX1 | 0.3 | 94.6 | 19.5 | 6 | 5 | 0.47 | 2.17 | No |
| 41 | CXCR4 | 8.9 | 59.4 | 35 | — | — | — | — | No |
| 42 | CXCR4 | 0.9 | 68.3 | 8.5 | 10 | 5 | 0.15 | 5.25 | No |
| 43 | CXCR4 | 5.1 | 1.9 | 19 | 8 | 5 | 0.10 | 7.39 | No |
| 44 | CXCR4 | 0.1 | 45.2 | 5 | 1 | 5 | 0.32 | 6.70 | No |
| 45 | CXCR4 | 7.3 | 3.7 | 22 | 1 | 5 | 0.92 | 1.36 | No |
| 46 | SBDS | 0.6 | 33.2 | 6.4 | — | — | — | — | No |
| 47 | SBDS | 5.1 | 26.2 | 15 | 10 | 7.5 | 0.42 | 22.16 | No |
| 48 | SBDS | 0.3 | 7.2 | 6 | — | — | — | — | No |
| 49 | SBDS | 1.7 | 4.1 | 9 | — | — | — | — | No |
| 50 | SBDS | 0.9 | 0.3 | 8.5 | 1 | 5 | 0.52 | 3.14 | No |
| 51 | SBDS | 0.3 | 1.4 | 5 | 4 | 5 | 0.36 | 1.32 | No |
| 52 | SBDS | 0.4 | 29.4 | 6 | — | — | — | — | No |
| 53 | SBDS | 0.6 | 0.8 | 7.8 | 1 | 5 | 0.37 | 2.39 | No |
| 54 | WAS | 6.4 | 6.5 | NA | — | — | — | — | No |
| 55 | G6PC3 | 0.8 | 13.1 | 8 | — | — | — | — | No |
| 56 | SPR54 | 3.1 | 2.9 | 14.5 | — | — | — | — | No |
| No. . | Mutated gene . | Age at first visit (y) . | Follow-up period (mo) . | Body weight (kg) . | G-CSF . | HSCT∗ . | |||
|---|---|---|---|---|---|---|---|---|---|
| Injection period (d) . | Dosage (μg/kg) . | bANC (×109 cells per L)† . | pANC (×109 cells per L)‡ . | ||||||
| 1 | ELANE | 13.5 | 1.6 | 33.5 | 9 | 7 | 0.53 | 2.37 | No |
| 2 | ELANE | 1.1 | 2.0 | 10.0 | 59 | 10-20 | 0.03 | 0.45 | No |
| 3 | ELANE | 1.8 | 20.8 | 12.5 | 14 | 10 | 0.65 | 2.01 | Yes |
| 4 | ELANE | 1.1 | 3.0 | 10 | — | — | — | — | No |
| 5 | ELANE(CyN) | 4.8 | 1.7 | 15 | — | — | — | — | No |
| 6 | ELANE | 1.4 | 2.3 | 11.5 | — | — | — | — | No |
| 7 | ELANE | 2.7 | 6.5 | 12 | 11 | 10 | 0.32 | 1.45 | Yes |
| 8 | ELANE | 1.4 | 2.4 | 9.5 | 9 | 5-10 | 0.34 | 2.41 | No |
| 9 | ELANE | 1.0 | 80.3 | 11.5 | 5 | 5-7.5 | 0.08 | 0.30 | No |
| 10 | ELANE | 0.2 | 8.1 | 6.6 | 15 | 7.5 | 0.47 | 5.01 | Yes |
| 11 | ELANE(CyN) | 6.7 | 0.5 | 23 | — | — | — | — | No |
| 12 | ELANE | 0.2 | 113.4 | 10 | 7 | 5-20 | 0.24 | 1.37 | No |
| 13 | ELANE | 1.5 | 7.8 | NA | — | — | — | — | No |
| 14 | ELANE | 13.4 | 12.0 | NA | — | — | — | — | Yes |
| 15 | ELANE | 1.5 | 29.9 | 12.5 | 53 | 7.5-15 | 0.91 | 2.11 | Yes |
| 16 | ELANE(CyN) | 10.1 | 2.1 | 26 | 10 | 5 | 0.45 | 10.33 | No |
| 17 | ELANE | 3.1 | 33.1 | 18 | 3 | 5 | 0.40 | 2.57 | Yes |
| 18 | ELANE | 0.5 | 0.5 | 6 | 9 | 10-15 | 0.04 | 0.90 | No |
| 19 | ELANE | 9.1 | 0.6 | 32 | 3 | 5 | 0.68 | 4.17 | No |
| 20 | ELANE | 5.7 | 0.6 | 20 | 11 | 5-10 | 0.26 | 1.32 | No |
| 21 | ELANE(CyN) | 0.4 | 33.5 | 15 | 1 | 5 | 0.46 | 8.38 | No |
| 22 | ELANE | 0.4 | 11.4 | 8 | 9 | 5 | 0.33 | 1.94 | No |
| 23 | ELANE | 2.5 | 0.9 | 11.5 | — | — | — | — | No |
| 24 | ELANE | 1.4 | 0.3 | 10 | — | — | — | — | Yes |
| 25 | ELANE | 1.0 | 60.0 | 9 | 93 | 5-20 | 0.05 | 0.98 | Yes |
| 26 | ELANE | 5.0 | 51.4 | 17 | 3 | 5 | 0.40 | 2.24 | No |
| 27 | ELANE | 2.5 | 19.6 | 12 | 50 | 5-7.5 | 0.18 | 1.71 | Yes |
| 28 | ELANE | 0.5 | 12.3 | 6 | 3 | 5 | 0.96 | 1.30 | No |
| 29 | ELANE | 0.8 | 1.5 | 8 | 4 | 5 | 0.90 | 1.07 | No |
| 30 | ELANE | 1.8 | 38.5 | 10 | 46 | 5-7.5 | 0.55 | 0.92 | Yes |
| 31 | ELANE | 6.5 | 15.4 | 16 | 9 | 5 | 0.42 | 0.55 | Yes |
| 32 | ELANE | 3.0 | 1.6 | 13 | 1 | 5 | 0.20 | — | No |
| 33 | ELANE | 1.1 | 1.4 | 10 | 10 | 5-10 | 0.11 | 3.81 | Yes |
| 34 | ELANE | 13.2 | 0.6 | 12 | 6 | 5-10 | 0.43 | 1.34 | No |
| 35 | ELANE | 6.6 | 3.3 | 22 | 7 | 5-10 | 0.18 | 2.37 | Yes |
| 36 | ELANE | 0.1 | 15.3 | 3 | 7 | 10-15 | 0.08 | 1.50 | No |
| 37 | HAX1 | 1.0 | 13.7 | 10 | 9 | 5-15 | 0.06 | 1.19 | No |
| 38 | HAX1 | 3.7 | 1.2 | 11 | 2 | 5 | 0.03 | 0.51 | No |
| 39 | HAX1 | 10.9 | 56.5 | 31 | 5 | 5 | 0.22 | 1.68 | No |
| 40 | HAX1 | 0.3 | 94.6 | 19.5 | 6 | 5 | 0.47 | 2.17 | No |
| 41 | CXCR4 | 8.9 | 59.4 | 35 | — | — | — | — | No |
| 42 | CXCR4 | 0.9 | 68.3 | 8.5 | 10 | 5 | 0.15 | 5.25 | No |
| 43 | CXCR4 | 5.1 | 1.9 | 19 | 8 | 5 | 0.10 | 7.39 | No |
| 44 | CXCR4 | 0.1 | 45.2 | 5 | 1 | 5 | 0.32 | 6.70 | No |
| 45 | CXCR4 | 7.3 | 3.7 | 22 | 1 | 5 | 0.92 | 1.36 | No |
| 46 | SBDS | 0.6 | 33.2 | 6.4 | — | — | — | — | No |
| 47 | SBDS | 5.1 | 26.2 | 15 | 10 | 7.5 | 0.42 | 22.16 | No |
| 48 | SBDS | 0.3 | 7.2 | 6 | — | — | — | — | No |
| 49 | SBDS | 1.7 | 4.1 | 9 | — | — | — | — | No |
| 50 | SBDS | 0.9 | 0.3 | 8.5 | 1 | 5 | 0.52 | 3.14 | No |
| 51 | SBDS | 0.3 | 1.4 | 5 | 4 | 5 | 0.36 | 1.32 | No |
| 52 | SBDS | 0.4 | 29.4 | 6 | — | — | — | — | No |
| 53 | SBDS | 0.6 | 0.8 | 7.8 | 1 | 5 | 0.37 | 2.39 | No |
| 54 | WAS | 6.4 | 6.5 | NA | — | — | — | — | No |
| 55 | G6PC3 | 0.8 | 13.1 | 8 | — | — | — | — | No |
| 56 | SPR54 | 3.1 | 2.9 | 14.5 | — | — | — | — | No |
—, not receiving G-CSF treatment.
HSCT, hematopoietic stem cell transplantation.
bANC, last absolute baseline neutrophil count (×109 cells per L) before G-CSF initiation.
pANC, peak ANC (×109 cells per L) within 10 days following G-CSF initiation.
Relationship between BM neutrophil lineage development and G-CSF responsiveness in different patient groups. (A) bANC before G-CSF treatment and pANC after G-CSF treatment in 36 patients who underwent concurrent G-CSF therapy and ANC monitoring. pANC, peak ANC levels within 10 days after the initiation of G-CSF. Student t test was used for comparation of ANC before and after G-CSF treatment within each group. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Bars represent means ± SD. (B) Statistical analysis of the correlation between the proportion of BM neutrophilic lineage cells and peripheral pANC, using Spearman correlation test. (C) Detailed analysis of the percentage of BM neutrophilic lineage cells and peripheral pANC in 30 patients who received G-CSF treatment concomitant with a cell morphology assay of BM smears. (D) Correlation between pANC and the populations of stem cell pool (myeloblast), the mitotic cell pool (promyelocyte, myelocyte) and the postmitotic cell pool (metamyelocyte, band cell and PMN), following the criteria described by Hong et al,24 using Spearman correlation test.
Relationship between BM neutrophil lineage development and G-CSF responsiveness in different patient groups. (A) bANC before G-CSF treatment and pANC after G-CSF treatment in 36 patients who underwent concurrent G-CSF therapy and ANC monitoring. pANC, peak ANC levels within 10 days after the initiation of G-CSF. Student t test was used for comparation of ANC before and after G-CSF treatment within each group. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Bars represent means ± SD. (B) Statistical analysis of the correlation between the proportion of BM neutrophilic lineage cells and peripheral pANC, using Spearman correlation test. (C) Detailed analysis of the percentage of BM neutrophilic lineage cells and peripheral pANC in 30 patients who received G-CSF treatment concomitant with a cell morphology assay of BM smears. (D) Correlation between pANC and the populations of stem cell pool (myeloblast), the mitotic cell pool (promyelocyte, myelocyte) and the postmitotic cell pool (metamyelocyte, band cell and PMN), following the criteria described by Hong et al,24 using Spearman correlation test.
ANC after G-CSF administration in different CN groups
| CN group . | G-CSF responsiveness . | Total cases (n) . | |||
|---|---|---|---|---|---|
| pANC ≥3.0 . | 1.5 ≤ pANC<3.0 . | 1.0 ≤ pANC<1.5 . | pANC<1.0 . | ||
| SCN1 n (%) | 3 (12%) | 10 (40%) | 6 (24%) | 6 (24%) | 25 |
| SCN3 n (%) | 0 (0%) | 2 (50%) | 1 (50%) | 1 (50%) | 4 |
| CyN n (%) | 2 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 |
| WHIM n (%) | 3 (75%) | 0 (0%) | 1 (50%) | 0 (0%) | 4 |
| SBDS n (%) | 2 (50%) | 1 (25%) | 1 (25%) | 0 (0%) | 4 |
| CN group . | G-CSF responsiveness . | Total cases (n) . | |||
|---|---|---|---|---|---|
| pANC ≥3.0 . | 1.5 ≤ pANC<3.0 . | 1.0 ≤ pANC<1.5 . | pANC<1.0 . | ||
| SCN1 n (%) | 3 (12%) | 10 (40%) | 6 (24%) | 6 (24%) | 25 |
| SCN3 n (%) | 0 (0%) | 2 (50%) | 1 (50%) | 1 (50%) | 4 |
| CyN n (%) | 2 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 |
| WHIM n (%) | 3 (75%) | 0 (0%) | 1 (50%) | 0 (0%) | 4 |
| SBDS n (%) | 2 (50%) | 1 (25%) | 1 (25%) | 0 (0%) | 4 |
pANC, peak ANC (×109 cells per L) within 10 days after G-CSF initiation.
To investigate the relationship between G-CSF responsiveness and BM neutrophil development in different patient groups, we statistically analyzed the correlation between the development of the neutrophilic lineage in BM and pANC. We observed a significant correlation that patients with poor BM neutrophil development exhibited reduced responsiveness to G-CSF (Figure 3B). Specifically, SCN1 and SCN3, which had lower percentages of BM neutrophilic lineage cells, also showed lower pANC after G-CSF administration (Figure 3C). In contrast, CyN, WHIM, and SBDS, which had higher percentages of BM neutrophilic cells, showed higher pANC. This suggests a reverse relationship between the degree of disruption in granulocyte development and the effectiveness of G-CSF treatment.
We further categorized BM neutrophil lineage cells into the stem cell pool (myeloblast), the mitotic cell pool (promyelocyte and myelocyte) and the postmitotic cell pool (metamyelocyte, band cell, and PMN) as described.24 Our analysis revealed a significant correlation between pANC and the populations of both mitotic and postmitotic pools (Figure 3D). However, no such correlation was observed between pANC and the stem cell pool (Figure 3D). This finding highlights the role of G-CSF in promoting early granulocyte mitosis and mobilizing cells from the postmitotic pool, and its relation to the increase in pANC.
We also conducted a Spearman analysis to assess the relationship between bANC before G-CSF treatment and the pANC after treatment. Our analysis did not reveal a significant correlation between these 2 measures (supplemental Figure 2). It appears that bANC does not predict responsiveness to G-CSF treatment.
Elevated serum Ig levels in SCN1, SCN3, and CyN correlate with increased BM plasma cells and serum APRIL concentrations
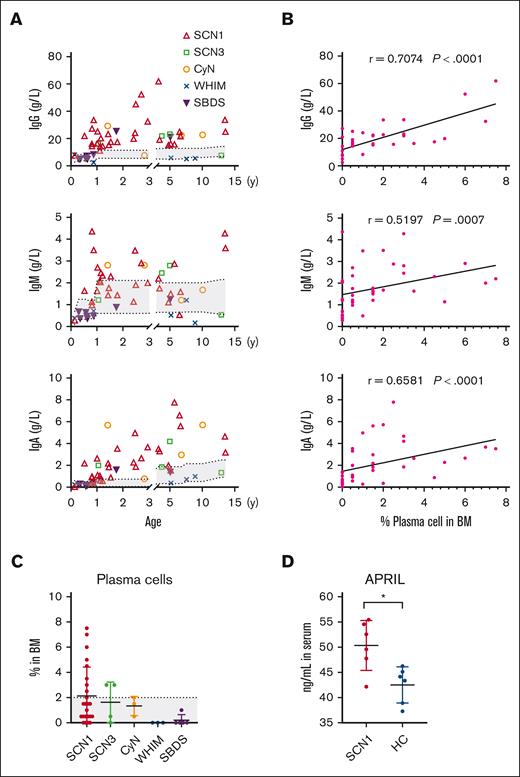
Adaptive immunity in patients with CN, who are characterized primarily by a neutrophil-mediated immune deficiency, has received little attention. We found that serum levels of IgG, IgM, and IgA were significantly elevated in the SCN1, SCN3, and CyN compared with those in the WHIM and SBDS, with SCN1 exceeding the reference range (Figure 4A). We also found a strong correlation between Ig levels and the percentages of BM PC cells in individual patients (Figure 4B). Analysis of lymphocyte subpopulations revealed diminished CD3+ T, CD4+ T, CD8+ T, and CD19+ B cells in patients in the WHIM group because of their impaired migration (supplemental Figure 3). Serum Igs are primarily secreted by long-lived plasma cells in the BM, which adhere to niches formed by BM stromal cells via CXCL12 and survive for decades by interacting with APRIL and interleukin-6.25 Consistently, we found a higher proportion of BM plasma cells in SCN1, SCN3, and CyN than in WHIM and SBDS (Figure 4C). In addition, serum APRIL concentrations were higher in patients in SCN1 group than those in age-matched healthy controls (Figure 4D). However, recent infections could influence Ig levels. Therefore, further studies are needed to elucidate the reasons behind the elevated Ig levels in SCN1, SCN3, and CyN.
SCN hyperimmunoglobulinemia is associated with elevated plasma cell counts and APRIL levels. (A) Serum Igs levels were measured by immunoturbidimetry in the patients. (B) Correlation between serum levels of each Ig isotype and BM PC in each patient, using Spearman correlation test. (C) Proportion of plasma cells in the BM, determined by cell morphology assays. Kruskal-Wallis test was applied to evaluate differences among 5 groups. (D) Serum APRIL levels in patients with SCN1 and age-matched healthy control (HC), measured by enzyme-linked immunosorbent assay. The gray area indicates normal reference range for each parameter. The Mann-Whitney test was used for SCN1 and HC comparison; ∗P < .05. Bars represent means ± SD.
SCN hyperimmunoglobulinemia is associated with elevated plasma cell counts and APRIL levels. (A) Serum Igs levels were measured by immunoturbidimetry in the patients. (B) Correlation between serum levels of each Ig isotype and BM PC in each patient, using Spearman correlation test. (C) Proportion of plasma cells in the BM, determined by cell morphology assays. Kruskal-Wallis test was applied to evaluate differences among 5 groups. (D) Serum APRIL levels in patients with SCN1 and age-matched healthy control (HC), measured by enzyme-linked immunosorbent assay. The gray area indicates normal reference range for each parameter. The Mann-Whitney test was used for SCN1 and HC comparison; ∗P < .05. Bars represent means ± SD.
Discussion
In this study, we conducted a comprehensive, retrospective systematic analysis of 56 patients with CN with 7 identified genes. Notably, we observed regional variations in the population distribution of CN pathogenic genes. Specifically, CXCR4 mutations were more prevalent in mainland China, whereas CLPB and G6PT mutations, commonly found in other regions,8,26 were not frequent in China. The overall infection characteristics of patients were similar across different CN groups, indicating that the reduction in neutrophil numbers resulting from different genetic variants was the dominant cause of infections in patients with CN. The higher occurrence of oral ulcers or skin infections in the SCN1 may result from a higher incidence of bacterial or fungal infections. On the other hand, the higher incidence of gastrointestinal symptoms (including diarrhea) in the SBDS group may be a consequence of exocrine pancreatic dysfunction.27
Our findings revealed that the development of neutrophils in the BM was affected much earlier and more severely in both SCN1 and SCN3 than in the CyN, WHIM, and SBDS groups, which was generally consistent with the number of neutrophils in the peripheral blood circulation. This suggests that patients in SCN1 and SCN3 group have poor BM neutrophil development, directly leading to severely reduced peripheral neutrophils. In contrast, patients in CyN and SBDS group have less severely affected BM neutrophil development, and consequently have higher neutrophil counts in the peripheral blood. Although BM neutrophil development is normal in WHIM, these patients have low circulating neutrophils because of the inability of BM neutrophils to be released into the periphery. Moreover, SCN1 mainly affects neutrophil development with little impact on eosinophils, presumably because of the specific expression of ELANE in the neutrophilic granulocyte lineage in BM.28
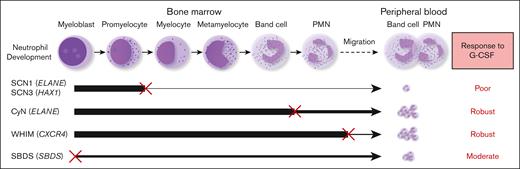
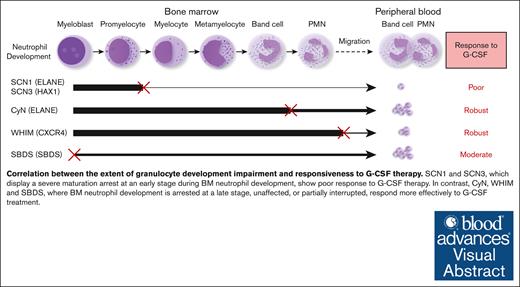
G-CSF is widely used in the treatment of CN and has achieved remarkable therapeutic outcomes.29 However, there is a lack of studies comparing the efficacy of different CN types to G-CSF administration, because patient responsiveness to G-CSF treatment has not been well-quantified. Dale et al compared the G-CSF responsiveness among CN, CyN, and autoimmune or idiopathic groups.20 They observed that the CN group had the lowest pre–G-CSF neutrophil counts. Upon G-CSF treatment, both the mean and median ANC increased in all groups. Our data are consistent with their findings and further elucidate the relationship between G-CSF responsiveness and granulocyte development. We found that the efficacy of G-CSF treatment inversely correlated with the extent of BM granulocyte development disruption in the patients. Our study reveals that different types of patients with CN have distinct responsiveness to G-CSF; SCN1 and SCN3, which display a maturation arrest at an early stage during BM neutrophil development, show poor response to G-CSF, whereas CyN, WHIM, and SBDS, in which BM neutrophil development is either disrupted at a late stage, unaffected, or moderately disrupted, responded more effectively to G-CSF therapy. This corresponds to the degress of their neutrophil development impairment (as detailed in Figure 5). Our findings establish a clear link between the severity of granulocyte development disruption and responsiveness to G-CSF therapy.
Correlation between the stage and severity of neutrophil development impairment and responsiveness to G-CSF therapy. SCN1 and SCN3, which display a severe maturation arrest at an early stage (between promyelocyte and myelocyte) during BM neutrophil development, show a poor response to G-CSF. CyN, WHIM, and SBDS, where BM neutrophil development is disrupted at a late stage, unaffected, or partially interrupted, respond well to G-CSF therapy. An ‘×’ indicates a developmental block or migration defect; The line thickness reflects the degree of normal neutrophil development.
Correlation between the stage and severity of neutrophil development impairment and responsiveness to G-CSF therapy. SCN1 and SCN3, which display a severe maturation arrest at an early stage (between promyelocyte and myelocyte) during BM neutrophil development, show a poor response to G-CSF. CyN, WHIM, and SBDS, where BM neutrophil development is disrupted at a late stage, unaffected, or partially interrupted, respond well to G-CSF therapy. An ‘×’ indicates a developmental block or migration defect; The line thickness reflects the degree of normal neutrophil development.
Although hypergammaglobulinemia has been described in patients with SCN1, the underlying mechanisms are not clear.3 A few studies have suggested an association between neutrophil counts, their elastase activity, and humoral immunity. For example, neutrophil numbers have been shown to negatively correlate with IgA levels.30 More recently, ELANE protein inhibitors were shown to stimulate mouse B cells to differentiate into plasma cells that produce IgG or IgA, and can also upregulate the transcription of AID, interleukin-10, BAFF, and APRIL coding genes.31 We observed that Ig levels and the proportion of lymphocyte subsets varied among patients with CN. Specifically, we noted elevated serum levels of IgG, IgA, and IgM and increased BM plasma cells in SCN1 caused by ELANE mutations. In addition, a higher serum APRIL concentration was found in SCN1 compared with healthy controls. Our results suggest that the increase in serum Ig levels in SCN1 is a result of an increased number of BM plasma cells, attributable to factors, such as APRIL, which promotes the survival of B cells and plasma cells. Although serum Ig levels were significantly increased in the ELANE group, such Igs with normal physiological functions do not cause disease manifestations, such as autoimmunity in patients. Instead, they may enhance patients’ humoral immune responses.
Conclusions
Clinical presentations and immunophenotypes displayed considerable variability among the different types of CN. SCN1 and SCN3 were more susceptible to skin infections, oral ulcers, and fungal infections. They also exhibited lower counts of peripheral neutrophils, a severe block of early neutrophil development in BM, elevated serum Ig levels, and a poor response to G-CSF therapy. CyN presented with moderately low peripheral neutrophil counts, well-developed BM granulocytes, normal serum Ig levels, and good responsiveness to G-CSF administration. Patients with WHIM, while displaying lower counts of peripheral WBCs, granulocytes, lymphocytes, and monocytes, along with lower Ig levels, presented with normal BM neutrophil development and responded well to G-CSF treatment. Patients with SBDS were more prone to diarrhea and exhibited moderately low peripheral neutrophil counts, partial impairment in BM neutrophil development, reduced Ig levels, and positive response to G-CSF therapy.
Acknowledgments
The authors thank the clinicians in the Department of Clinical Immunology, Children’s Hospital of Fudan University and the members in Wang laboratory at the Department of Immunology of School of Basic Medical Sciences for their helpful discussions.
This work was supported by the Major Research Plan of the National Natural Science Foundation of China (grant 32330033; J.-Y.W.), the National Natural Science Foundation of China (grant 32270932; J.-Y.W.), Projects of International Cooperation and Exchanges NSFC (grant 82011540008; J.-Y.W.), Shanghai Municipal Science and Technology Major Project (grant ZD2021CY001; X.W.), National Natural Science Foundation of China for Young Scholar (grant 82202013; Q.M.), and China Postdoctoral Science Foundation Grant (grant 2022M720782; Q.M.).
Authorship
Contribution: X.M. and J.H. designed the study, and collected and analyzed the clinical and immunological data; X.M. provided a draft of the manuscript; H.Z., L.L., and L.D. participated in data collection and analysis; Q.M. provided suggestions for the criteria for grouping; Y.L. provided support in healthy control samples and flow cytometry; J.H., L.L., W.W., W.Y., J.S., and X.W. diagnosed and treated patients and helped collect the clinical data of the patients; Q.M., H.Z., L.D., Y.L., and M.Y. corrected the manuscript; X.W. and J.-Y.W. supervised the study; and J.-Y.W. reviewed and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ji-Yang Wang, Department of Immunology, Fudan University, Room 921, West #13 Building, Dong'an Rd, Shanghai 200032, China; email: wang@fudan.edu.cn; Jia Hou, Department of Clinical Immunology, Children’s Hospital of Fudan University, National Children’s Medical Center, Fudan University, 399 Wanyuan Rd, Shanghai 201102, China; email: doctorhoujia@hotmail.com; and Xiaochuan Wang, Department of Clinical Immunology, Children’s Hospital of Fudan University, National Children’s Medical Center, Fudan University, 399 Wanyuan Rd, Shanghai 201102, China; email: xchwang@shmu.edu.cn.
References
Author notes
X.M. and H.Z. contributed equally to this work.
Additional data related to this study, as well as specific materials and protocols, can be obtained from the corresponding author, Ji-Yang Wang (wang@fudan.edu.cn) upon reasonable request.
The full-text version of this article contains a data supplement.

![Comparative analysis of BM or peripheral blood immune cell subsets across 5 groups using routine blood tests and cell morphology assays of BM smears. (A) Total number of WBC, granulocytes, monocytes, and lymphocytes in the peripheral blood. (B) Absolute numbers of granulocytes (neutrophils, eosinophils, and basophils) in the peripheral blood. The brown line in neutrophil indicates the cut-off value of 1.5 × 109 cells per L of neutropenia. (C) Proportion of granulocytes, monocytes, and lymphocytes in BM. (D) Proportion of specific granulocyte types (neutrophils, eosinophils, and basophils) in BM. (E-G) Proportions of BM neutrophilic (E-F) and eosinophilic (E,G) lineage cells at different developmental stages for each patient group. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Kruskal-Wallis test was used for comparison among 5 groups. Bars represent means ± standard deviation [SD]. The gray area indicates reference range of each index, as described in the Methods section.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/7/10.1182_bloodadvances.2023012171/2/m_blooda_adv-2023-012171-gr2.jpeg?Expires=1765057308&Signature=cRY2iKyEPnzpcTqbqBME08zekQEFYhUREfauIRvjerH6OsSZYElGvc0OCp83xn1a9ClTgPwCQB9G3LKprEu~ScwX1w3QCWfFbjcRyJnVYgqN-br545-79goADMNHJdkKemtKYuLC~o80GBVzFyDDi4vFDOVxggsR~IBk1P3qIGJQ7pw9rC34zM7~2QO5OphzJQauIc-3-fbSaL~fZzQ0JWIR7DLXsHivg1il0Qn6Xh8eqxflBRcAJCWABC3AL8foiJKweil9SC9BSkQWbmKDCt~PW469c1KvhgBrMRTYyQr7Upjp~Kf~jggaeIlO1tUUjO~pHDxAVkTLoXlW3h1v4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)