Key Points
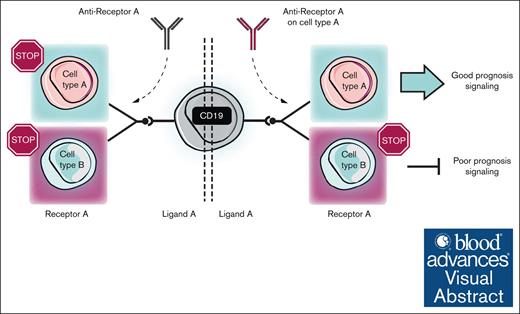
An identical molecular interaction can confer either good or poor prognosis, depending on the cell types involved.
Therapeutic strategies targeting ligand-receptor interactions should take into consideration the specific cell types involved.
Visual Abstract
Although most patients with multiple myeloma respond to treatment initially, therapy resistance develops almost invariably, and only a subset of patients show durable responses to immunomodulatory therapies. Although the immune microenvironment has been extensively studied in patients with myeloma, its composition is currently not used as prognostic markers in clinical routine. We hypothesized that the outcome of immune signaling pathway engagement can be highly variable, depending on which 2 cellular populations participate in this interaction. This would have important prognostic and therapeutic implications, suggesting that it is crucial for immune pathways to be targeted in a specific cellular context. To test this hypothesis, we investigated a cohort of 25 patients with newly diagnosed multiple myeloma. We examined the complex regulatory networks within the immune compartment and their impact on disease progression. Analysis of immune cell composition and expression profiles revealed significant differences in the B-cell compartment associated with treatment response. Transcriptional states in patients with short time to progression demonstrated an enrichment of pathways promoting B-cell differentiation and inflammatory responses, which may indicate immune dysfunction. Importantly, the analysis of molecular interactions within the immune microenvironment highlights the dual role of signaling pathways, which can either be associated with good or poor prognosis depending on the cell types involved. Our findings therefore argue that therapeutic strategies targeting ligand-receptor interactions should take into consideration the composition of the microenvironment and the specific cell types involved in molecular interactions.
Introduction
Tumors embody complex ecosystems, and variability in the composition of immune components and expression are linked to distinct cancer subtypes and treatment results. Despite initial therapeutic responses in patients with multiple myeloma (MM), treatment resistance often leads to eventual relapse. Immunomodulatory drugs (IMiDs), thalidomide and its derivatives lenalidomide and pomalidomide, have become pervasive in myeloma treatment1-3 due to their diverse mechanisms, including antiangiogenic, cytotoxic, and immunomodulatory properties.4,5 However, despite their efficacy,6-8 IMiDs do not yield lasting responses in all patients, and resistance is common. Beyond IMiDs, other immunotherapies have shown success in myeloma treatment, underlining the critical role of the bone marrow immune microenvironment.9 Nevertheless, persistent immunotherapeutic responses are observed only in a subset of patients. Therefore, to enhance patient outcomes, we need a better understanding of the immune microenvironment, the factors determining response to therapies, and predictive markers for response and disease progression.
The bone marrow immune microenvironment is composed of a large variety of immune cells, including CD4+ and CD8+ T cells, regulatory T cells (Tregs), myeloid-derived suppressor cells, natural killer (NK) cells, and diverse B-cell subsets. Past research indicates considerable shifts within the MM bone marrow microenvironment, such as cytokine secretion and compositional changes.10 MM cells have been shown to induce immunosuppression via elevated numbers of Tregs, myeloid-derived suppressor cells, nontraditional monocytes, and alterations in T- and NK-cell populations.10 Yet, the implications of changes in the B-cell compartment on the antitumor immune response or immune evasion remain undetermined.
Compositional shifts in the cellular architecture of the bone marrow play a vital role in the initiation and progression of MM.11,12 These shifts support proinflammatory states and immunosuppression, thereby enabling immune evasion and disease progression through intricate interactions between immune cells, stromal elements, and malignant plasma cells.11-13 In this study, we aim to better comprehend the complex interregulation within the immune compartment in patients with newly diagnosed (ND) MM, identify key participants, and their influence on treatment resistance and disease progression.
Importantly, the heterogeneity of immune cell composition among patients has potential as a critical biomarker, carrying prognostic value and influencing therapeutic strategies.10 Here, we correlated alterations in cell type composition and expression profiles with therapy response. Our findings delineate the transcriptional and compositional shifts in the bone marrow immune microenvironment during disease progression, highlighting mechanisms for antitumor immune response and immune evasion. We examined cell-cell interactions to identify the factors that condition the fate and functionality of immune cell subsets in the bone marrow. Interestingly, our studies of molecular interactions within immune compartments revealed that the same signaling pathway can convey either positive or negative prognosis, contingent on the specific cell types interacting. We propose a model underscoring the dual role of cell-cell interactions, in which the same interaction can generate both favorable and adverse outcomes depending on the involved cell types and signal directionality. Our studies support the notion that therapies aimed at targeting ligand-receptor interactions should consider the microenvironment's composition and the particular cell types partaking in molecular interactions to block harmful signals while preserving beneficial ones.
Methods
Patient samples
Cryopreserved CD138− bone marrow samples from 25 patients with treatment-naive ND myeloma (NDMM) were analyzed in this study (supplemental Table 1). Bone marrow from normal donors was obtained from AllCells.
Cell sorting
Cells were stained with antibodies against CD45 fluorescein isothiocyanate (CD45 FITC) (HI30, eBioscience; 1:100), CD3 peridinin-chlorophyll-protein complex-cyanine 5.5 (CD3 PerCP-Cy5.5) (OKT3, eBioscience; 1:100), CD14 allophycocyanin-cyanine 7 (CD14 APC-Cy7) (MoP9, BD Biosciences; 1:100), and CD19 phycoerythrin (CD19 PE) (HIB19, BioLegend; 1:100). Dead cells were excluded with DAPI (4′,6-diamidino-2-phenylindole) (1 μg/mL; Sigma-Aldrich). A total of 100 cells per well were sorted into 96-well plates using a Sony SH800 sorter. Percentages of positive cells were calculated using FlowJo software v10. Statistical analysis was performed using GraphPad software.
Low-input RNA-seq library preparation
Processing of RNA-seq data
Sequencing reads were trimmed using trimmomatic and aligned to the human genome (version hg19) using STAR aligner with following parameters “-- twopassMode Basic –alignIntronMax 100000 --alignMatesGapMax 100000 --alignSJDBoverhangMin 10 --alignSJstitchMismatchNmax 5 -1 5 5.”16,17 HTSeq18 and RSEM19 were used to obtain raw counts and normalized TPM (Transcripts Per Million) values from the aligned BAM files.
Quality control and processing of RNA-seq data
To remove low-quality samples from our data set, we used the following cutoffs: for library size, detected genes, and percentage counts mapping to ribosomal genes, 4 median-absolute-deviations; for mitochondrial genes, 2 median-absolute-deviations. Dimensionality reduction and clustering of high-quality samples were performed using the scater20 and scran packages.21 Signaling pathway activity was inferred using the PROGENy package.21,22
Deconvolution analysis
Cell type composition from bulk expression profiles was inferred using Cibersort.23 Cibersort was run using default settings, providing the normalized expression matrix as input. Statistical analysis was performed using GraphPad software.
Inference of cell-cell interactions
Cell-cell interactions were inferred based on ligand-receptor coexpression using the CellphoneDB package (v2.0) with default settings.24 Analysis of cell-cell interactions was performed separately for patients with good (time to progression [TTP] high) or poor (TTPlow) prognosis. Interactions were considered specific to 1 prognosis (“prognostic interactions”) if they were detected as significant (P < .05) in 1 condition but not the other (P ≥ .05).
Validation of gene expression
Validation cohorts
Published single-cell RNA-seq data from patients treated with IMiDs27-29 or non-IMiD–based treatments28,30 were obtained for validation. Data were preprocessed using Cell Ranger Ranger 6.1.031 and Seurat V4.15 Celltype annotation was performed using azimuth and the human bone marrow data set as a reference. CITE-seq (Cellular Indexing of Transcriptomes and Epitopes by sequencing) data were obtained from GSE210079.30
Informed consent has been obtained from all patients in this study.
Results
Heterogeneous bone marrow immune cell composition in patients with treatment-naive MM
We hypothesized that the outcome of activating an immune signaling pathway can vary based on the cell types involved in the interaction. Because immune cell composition is heterogeneous in MM,10 we first sought to identify those cell populations with the greatest difference in abundance between patients with better or worse outcomes, postulating that these would be most likely to engage in interactions that are relevant for patient outcome.
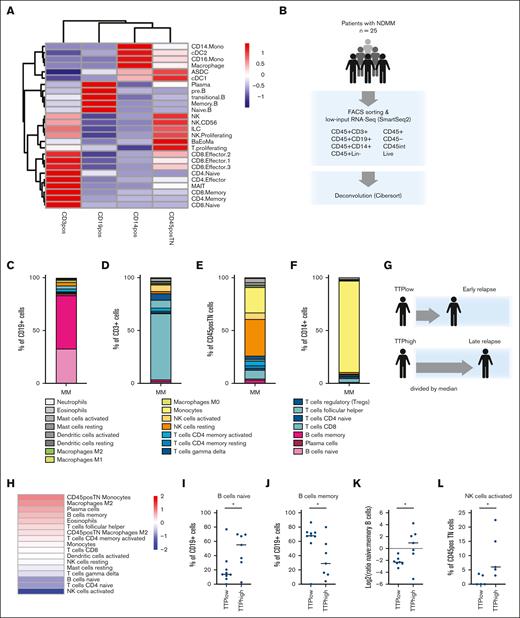
To define the composition of the bone marrow immune compartment in treatment-naive patients with MM, we investigated bone marrow aspirates from a cohort of 25 patients with NDMM. The median time to progression (TTP) in this cohort was 28 months, and 76% of patients received first-line treatment with an IMiD (Figure 1A-B). We performed fluorescence activated cell sorting (FACS) to enrich for distinct immune cell populations in the bone marrow, CD45+CD3+ cells to enrich for T cells, CD45+CD19+ to enrich for B cells, CD45+CD14+ to enrich for classical monocytes, and CD45+CD3−CD19−CD14− (CD45+Lin−) to enrich for other immune cell subtypes (Figure 1C; supplemental Figure 1A). Consistent with previous studies,10 patient bone marrow immune cell compositions demonstrated considerable variability, with CD3+ cells accounting for 1% to 77% of CD45+ immune cells in the bone marrow (mean, 30%; range, 1%-77%), CD19+ cells accounting for 0% to 22% (mean, 6%; range, 0%-22%), CD14+ for 3% to 56% (mean, 16%; range, 3%-57%), and CD45+Lin− for 7% to 85% (mean, 49%; range, 7%-85%; Figure 1D).
Characterization of bone marrow immune environment in patients with treatment-naive MM. (A-B) Patient characteristics. (A) Time to progression (TTP) for 25 patients with data availability (median, 28 months; range, 0-72 months). (B) Treatment data for patients included in this study. Shown is percentage of patients who received a first-line treatment including an IMiD. (C) Diagram showing sorting strategy. (D) Flow percentages for individual cell populations. Patients are ordered by TTP. (E) Patients with early (TTPlow) and late (TTPhigh) relapse were divided by median TTP. (F) Bar plot showing flow percentages for individual populations comparing TTPlow vs TTPhigh patients. Significance was assessed using an unpaired t test; ∗P ≤ .05. (G) Dot plots showing the distribution of frequencies for individual cell populations shown in panel F. Significance was assessed using an unpaired t test; ∗P ≤ .05. (H) Experimental workflow. Low-input RNA-seq of 100-cell pools was performed using the Smart-Seq2 protocol. (I) Tsne plot showing samples colored by sort marker. (J) Expression of canonical marker genes for individual cell types across immune cell subsets. Tsne, t-distributed stochastic neighbor embedding.
Characterization of bone marrow immune environment in patients with treatment-naive MM. (A-B) Patient characteristics. (A) Time to progression (TTP) for 25 patients with data availability (median, 28 months; range, 0-72 months). (B) Treatment data for patients included in this study. Shown is percentage of patients who received a first-line treatment including an IMiD. (C) Diagram showing sorting strategy. (D) Flow percentages for individual cell populations. Patients are ordered by TTP. (E) Patients with early (TTPlow) and late (TTPhigh) relapse were divided by median TTP. (F) Bar plot showing flow percentages for individual populations comparing TTPlow vs TTPhigh patients. Significance was assessed using an unpaired t test; ∗P ≤ .05. (G) Dot plots showing the distribution of frequencies for individual cell populations shown in panel F. Significance was assessed using an unpaired t test; ∗P ≤ .05. (H) Experimental workflow. Low-input RNA-seq of 100-cell pools was performed using the Smart-Seq2 protocol. (I) Tsne plot showing samples colored by sort marker. (J) Expression of canonical marker genes for individual cell types across immune cell subsets. Tsne, t-distributed stochastic neighbor embedding.
To assess whether bone marrow immune cell composition correlated with TTP, we divided the patient cohort by median TTP into TTPlow patients displaying early relapse and TTPhigh patients showing late relapse (Figure 1E). We compared the proportions of immune cell subsets between conditions and observed that patients with early relapse displayed higher proportions of CD14+ cells (P = .0295, by unpaired t test), arguing that the presence of monocytes is negatively correlated with TTP (Figure 1F-G). Because we observed a high degree of variability between patients, we also compared the 25th and 75th percentiles and observed that patients with very late relapse showed a higher proportion of CD3+ cells and CD19+ cells than patients with very early relapse, indicating that greater numbers of T and B lymphocytes have a favorable impact on prognosis (P = .03 and P = .005, by unpaired t test, respectively; supplemental Figure 1B-D).
To define the cell type composition and expression profiles of different immune cell subtypes in the bone marrow in greater detail, we next performed FACS and low-input RNA-seq using the Smartseq2 technology (Figure 1H). To enhance the detection of genes, improve data quality, and enhance statistical power, we implemented a strategy of examining “minipools” of cells. After quality control filtering, we performed clustering and visualization by t-distributed stochastic neighbor embedding as an initial assessment of heterogeneity (Figure 1I; supplemental Figure 2A-B). Clustering was determined by sorted surface marker but not by individual or prognostic conditions (Figure 1I; supplemental Figure 2C-G). We next assessed the expression of lineage-defining marker genes, including CD3D for T cells, CD19 for B cells, and CD14 for monocytes (supplemental Figure 2E-G). We confirmed this by assessing the expression of canonical marker genes for different immune cell types (Figure 1J).
Differential enrichment of B-cell subsets associated with prognosis
To define in greater detail which cell types and maturation states were enriched in the various immune compartments, we assessed the expression of gene signatures (Figure 2A). We recovered the expression of gene signatures for pre-B cells, naive B cells, transitional B cells, memory B cells, and plasma cells in the CD19+ compartment, indicating that this compartment contains a mixture of B-cell subtypes. Similarly, in the CD14+ compartment, we detected the expression of marker genes for monocytes, dendritic cells, and macrophages. In the CD3+ compartment, we detected the expression of gene signatures related to various CD8+ and CD4+ T-cell subtypes and, to a lesser degree, NK cells, presumably because both cell types share the expression of certain genes, whereas the CD45+CD3−CD19−CD14− compartment was enriched in NK cells and dendritic cells.
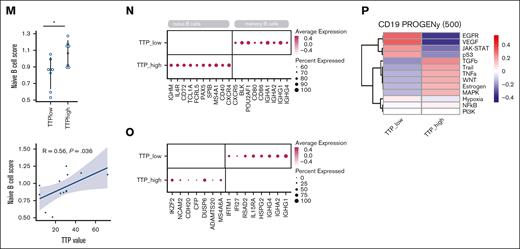
Differential enrichment of B-cell subsets associated with prognosis. (A) Analysis of the cell types composing the flow-sorted CD3+, CD14+, CD19+, and CD45+TN immune compartments according to gene signatures from azimuth.15 (B) Experimental workflow. Low-input RNA-seq was performed of individual cell populations sorted based on indicated markers from the bone marrow of 25 patients with NDMM. Cibersort was used to deconvolute cell type composition. (C-F) Bar graphs showing the composition of individual immune cell compartments based on deconvolution in patients with NDMM. Shown are percentages of cell types inferred by Cibersort for CD19+ cells (C), CD3+ cells (D), CD45+TN cells (E), and CD14+ cells (F). (G) Patients with early (TTPlow) and late (TTPhigh) relapse were divided by median TTP. (H) Heat map showing relative differences in cell type abundance between patients with poor vs good prognosis. (I-L) Dot plots showing the distribution of values for the indicated cell populations comparing TTPlow with TTPhigh. Significance was assessed using an unpaired t test; ∗P ≤ .05. (K) Log2 ratio of naive/memory B cells in TTPlow vs TTPhigh patients. Significance was assessed using an unpaired t test; ∗P ≤ .05. (M) Relative scores for a gene set for naive B cells in TTPlow vs TTPhigh patients. ∗P ≤ .05, by unpaired t test. Correlation of relative scores (bottom) for naive B-cell gene set vs TTP value (R = 0.56; P = .036). (N) Expression of canonical marker genes for naive and memory B cells in TTPlow vs TTPhigh patients. (O) Expression of marker genes distinguishing B cells in TTPlow and TTPhigh patients. (P) Heat map comparing activity of signaling pathways between TTPlow vs TTPhigh patients. ASDC, AXL+ dendritic cell; BaEoMa, basophil eosinophil mast progenitor; cDC1, CD141+ myeloid dendritic cell; cDC2, CD1c+ myeloid dendritic cell; CD8 effector.1, memory-like CD8+GZMK+ alpha-beta T cell; CD8 effector.2, late differentiated CD8+GZMB+ cytotoxic alpha-beta T cell; CD8 effector.3, NKT-like CD8+ alpha-beta T cell; ILC, innate lymphoid cell; MAIT, mucosal associated invariant T; Mono, monocyte; NKdim, CD56dim NK cell; NKbright, CD56bright NK cell.
Differential enrichment of B-cell subsets associated with prognosis. (A) Analysis of the cell types composing the flow-sorted CD3+, CD14+, CD19+, and CD45+TN immune compartments according to gene signatures from azimuth.15 (B) Experimental workflow. Low-input RNA-seq was performed of individual cell populations sorted based on indicated markers from the bone marrow of 25 patients with NDMM. Cibersort was used to deconvolute cell type composition. (C-F) Bar graphs showing the composition of individual immune cell compartments based on deconvolution in patients with NDMM. Shown are percentages of cell types inferred by Cibersort for CD19+ cells (C), CD3+ cells (D), CD45+TN cells (E), and CD14+ cells (F). (G) Patients with early (TTPlow) and late (TTPhigh) relapse were divided by median TTP. (H) Heat map showing relative differences in cell type abundance between patients with poor vs good prognosis. (I-L) Dot plots showing the distribution of values for the indicated cell populations comparing TTPlow with TTPhigh. Significance was assessed using an unpaired t test; ∗P ≤ .05. (K) Log2 ratio of naive/memory B cells in TTPlow vs TTPhigh patients. Significance was assessed using an unpaired t test; ∗P ≤ .05. (M) Relative scores for a gene set for naive B cells in TTPlow vs TTPhigh patients. ∗P ≤ .05, by unpaired t test. Correlation of relative scores (bottom) for naive B-cell gene set vs TTP value (R = 0.56; P = .036). (N) Expression of canonical marker genes for naive and memory B cells in TTPlow vs TTPhigh patients. (O) Expression of marker genes distinguishing B cells in TTPlow and TTPhigh patients. (P) Heat map comparing activity of signaling pathways between TTPlow vs TTPhigh patients. ASDC, AXL+ dendritic cell; BaEoMa, basophil eosinophil mast progenitor; cDC1, CD141+ myeloid dendritic cell; cDC2, CD1c+ myeloid dendritic cell; CD8 effector.1, memory-like CD8+GZMK+ alpha-beta T cell; CD8 effector.2, late differentiated CD8+GZMB+ cytotoxic alpha-beta T cell; CD8 effector.3, NKT-like CD8+ alpha-beta T cell; ILC, innate lymphoid cell; MAIT, mucosal associated invariant T; Mono, monocyte; NKdim, CD56dim NK cell; NKbright, CD56bright NK cell.
To comprehensively assess the relative abundance of different cell types in the bone marrow, we performed deconvolution of cell types using cibersort23 (Figure 2B-F). Consistent with our signature-based analysis, we identified naive and memory B cells as well as plasma cells in the CD19+ subset, CD4+ and CD8+ T cells in the CD3+ compartment, activated and resting NK cells as well as dendritic cells and presumably nonclassical monocytes in the CD45+TN (CD45+ triple negative/CD45+Lin-) subset, and mostly monocytes in the CD14+ subset.
We then assessed how cell type composition in patients with NDMM differed from that of normal donors (supplemental Figure 3A-C). Memory B cells were significantly enriched in patients with myeloma (P = .0106, by 2-tailed t test; mean of CD19+ cells in MM, 50.3%; range, 0%-83%; vs mean in ND, 8%; range, 0%-23%), as were T follicular helper (Tfh) cells (P = .046; mean of CD3+ cells in MM, 7.5%; range, 1.9%-18.9%; vs mean in ND, 2.7%; range, 0%-6.7%; supplemental Figure 3D). Tfh cells are commonly located in secondary lymphoid organs, including the tonsil, spleen, and lymph nodes, but they have been described in the bone marrow in hematological diseases.32,33 Interestingly, Tfh cells may help naive B cells to differentiate into memory B cells,34 which is consistent with our observation of differentiation toward a memory B-cell phenotype in MM. Conversely, we observed a decreased abundance of naive B cells (P = .0436, by t test; mean of CD19+ cells in MM, 32.7%; range, 0%-77%; vs mean in ND, 62.3%; range, 52%-71%) and M2 macrophages in MM (P = .0017, by t test; mean in MM, 1.4%; range, 0%-3.8%; vs mean in ND, 12.1%; range, 0.7%-19.7%).
To determine which of those cell types correlate with response to therapy, we compared cell type abundance in patients with early relapse vs late relapse (Figure 2G-L). Interestingly, the proportion of naive B cells was higher in patients with late relapse (P = .047, by t test; mean in TTPhigh patients, 47.5%; range, 7%-77%; vs mean in TTPlow patients, 21.2%; range, 2%-70%; Figure 2I), whereas the proportion of memory B cells was decreased in patients with late relapse than those with early relapse (P = .0488, by t test; mean in TTPhigh patients, 34.6%; range, 4%-80%; vs mean in TTPlow patients, 62.6%; range, 0%-87%; Figure 2J). The ratio of naive to memory B cells was significantly higher in TTPhigh patients (P = .041, by t test; Figure 2K), indicating that there is a shift from naive to memory B cells in patients with early relapse, and having less differentiated B cells is correlated with good prognosis. We also observed an increased abundance of activated NK cells in patients with late relapse (P = .0435, by t test; mean in TTPhigh patients, 10.4%; range, 3%-23%; vs mean in TTPlow patients, 1.3%; range, 0%-4%; Figure 2l). In line with previous studies,35 these data suggested that an immunocompetent microenvironment with adequate B-cell capacity is an important factor for the efficacy of IMiD-based therapy.
To define the functional effect of compositional changes in the bone marrow microenvironment between patients with short and long TTPs, we assessed gene expression signatures. We observed increased expression of a gene set associated with naive B cells in patients with late relapse (TTP high; P = .027; Figure 2M), consistent with our previous observations. We confirmed this association in an independent data set (GSE161801)28 consisting of 9 patients with relapsed/refractory (R/R) myeloma receiving IMiD-based treatments (P < 2.2e−16, by Wilcoxon test; supplemental Figure 4A). Furthermore, the expression score for naive B cells correlated with TTP (R = 0.56; P = .036).
When assessing the expression of marker genes for either naive or memory B cells, we detected enrichment in patients with late and early relapses, respectively (Figure 2N). Other marker genes in the CD19+ immune compartment in patients with late relapse included the transcription factor HELIOS (IKZF2), whereas in TTPlow patients, we detected expression of interferon-response genes, suggesting that the B-cell compartment might receive divergent signals from the environment (Figure 2O). We therefore analyzed cell signaling pathways (Figure 2P) using PROGENy.22 Consistent with the greater expression of interferon-responsive genes, we predicted greater activation of JAK-STAT signaling in patients with poor prognosis, as well as greater activity in EGFR (Epidermal Growth Factor Receptor) and VEGF (Vascular Endothelial Growth Factor) signaling pathways. In contrast, patients with good prognosis showed greater activation of the tumor necrosis factor α (TNF-α) and MAPK (Mitogen-Activated Protein Kinase) signaling pathways.
Prognostic impact of cell-cell interactions depends on involved cell types
Our data and prior studies indicate that the bone marrow microenvironment in MM exhibits dysregulation in receptor signaling,36 cytokine expression,28,36 and numerical alterations in immune cell subsets.10,37,38 Surface interactions are the main drivers of intracellular signaling and therefore control the fate and function of cells. To define the underlying causes that are conditioning the fate and functions of immune cell subsets in the bone marrow, we therefore investigated cell-cell interactions using CellPhoneDB.24
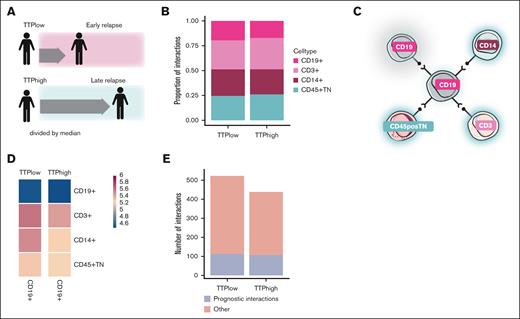
We predicted cell-cell interactions between and within the immune cell compartments in both prognostic conditions (Figure 3A-B). We found that the distribution of interactions of the different immune cell compartments was similar in both conditions, with slightly more interactions detected in the CD3+ compartment and less in the CD19+ compartment in TTPhigh patients (32% vs 29% of interactions for CD3+ and 17% vs 20% of interactions for CD19+ in TTPhigh vs TTPlow, respectively). Because we had identified significant compositional changes in the B-cell compartment, we next went on to assess cell interactions with a focus on CD19+ cells (Figure 3C-D). We identified a total of 438 inferred interactions in TTPhigh and 523 interactions in TTPlow patients (supplemental Tables 2 and 3). We observed that B cells interact the most with CD3+ cells and least with other CD19+ cells. In TTPlow patients, we observed more interactions with CD14+ cells than in TTPhigh patients (Figure 3D). To assess the association of these cell-cell interactions with prognosis in greater detail, we next filtered the interactions that were significant (P < .05) in 1 condition (TTP status), but those that were not significant (P ≥ .05) in the other TTP status (see “Methods”). These prognostic interactions accounted for 24% and 22% of total identified interactions in TTPhigh (107/438 [24%]) and TTPlow patients (113/523 [22%]), respectively (Figure 3E; supplemental Tables 2-5).
Association of the B-cell interactome with MM progression. (A) Diagram. Interactions were determined using CellPhoneDB24 for patients with early or late relapse. (B) Comparison of the number of CellPhoneDB-predicted interactions between TTPlow and TTPhigh samples. (C) Diagram illustrating B-cell–centric interactions in which B cells interact with various other cell types. (D) Heat map comparing the interactome of B cells with other cell types in TTPlow vs TTPhigh patients. (E) Number of prognostic interactions vs total interactions by condition.
Association of the B-cell interactome with MM progression. (A) Diagram. Interactions were determined using CellPhoneDB24 for patients with early or late relapse. (B) Comparison of the number of CellPhoneDB-predicted interactions between TTPlow and TTPhigh samples. (C) Diagram illustrating B-cell–centric interactions in which B cells interact with various other cell types. (D) Heat map comparing the interactome of B cells with other cell types in TTPlow vs TTPhigh patients. (E) Number of prognostic interactions vs total interactions by condition.
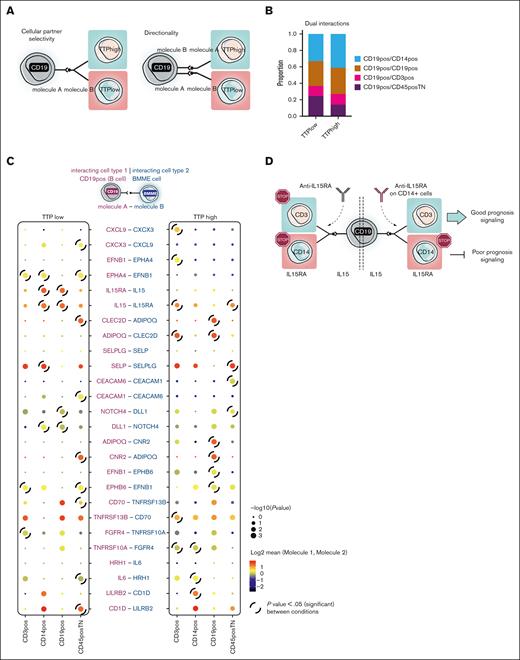
Remarkably, we observed that some of these “prognostic” interactions were present in both conditions at the same time. These interactions took place either between different cell types, or the ligand/receptor expression was switched between the involved cell types (illustrated in Figure 4A). These data suggest a model in which the selectivity of these dual interactions is determined by the cellular partner (cellular-partner selectivity) that the B cell interacts with. Thus, if the interacting receptor/ligand is expressed by cell type A, the interaction confers good prognosis, whereas the same interacting ligand/receptor expressed by cell type B confers poor prognosis. Alternatively, it is also conceivable that B cell expresses both the receptor and the ligand, and the prognosis is determined by the directionality of the signal, that is, the other cell type that expresses the ligand or the receptor. We referred to these interactions as “dual interactions” to highlight their ambiguous role, because the net effect on the prognosis is neither solely favorable nor unfavorable. We identified a total of 64 dual interactions (supplemental Table 6) and assessed their distribution across the different interacting cell types within each prognostic condition (Figure 4B). We observed an enrichment of these interactions in CD45+TN and CD3+ cells in the TTPhigh group and in CD14+ and CD19+ cells in the TTPlow group.
Dual interactions in B cells. (A) Model of dual interactions. Model of “Cellular-partner selectivity” (left). Given molecule A and molecule B (undefined ligand-receptor interacting pair), B cells (illustrated as CD19, gray) express exclusively molecule A on the cell surface and the other cells in their vicinity present molecule B. Prognosis is based on the engaged cell type. Model based on “directionality” of the signaling (right). Given molecule A and molecule B (undefined ligand-receptor interacting pair), the B cell (illustrated as CD19, gray) presents both ligand and receptor molecules. Prognosis is determined by the directionality of the signal upon binding of the complementary molecule, which is expressed by other cells in the surroundings. (B) Proportion of dual interactions established between each possible immune pair under study and their distribution in each prognostic condition. (C) Heat map showing dual interactions with potential targetability in MM. Displayed are the percentage of cells expressing the molecule and the median expression of both molecules by the pair of engaged cell types. Highlighted are those interactions which are significant (P < .05) in 1 condition but not the other. In the shown interactions, the first molecule is expressed in all cases by CD19+ cells (B cells); the second molecule is expressed by the 4 cell types (CD3+, CD14+, CD19+, or CD45+TN). On the left side of the list of interactions, the column represents TTPlow patients; on the right side, the column represents TTPhigh patients. (D) Model illustrating IL-15–IL-15RA interactions on diverse cell types as example.
Dual interactions in B cells. (A) Model of dual interactions. Model of “Cellular-partner selectivity” (left). Given molecule A and molecule B (undefined ligand-receptor interacting pair), B cells (illustrated as CD19, gray) express exclusively molecule A on the cell surface and the other cells in their vicinity present molecule B. Prognosis is based on the engaged cell type. Model based on “directionality” of the signaling (right). Given molecule A and molecule B (undefined ligand-receptor interacting pair), the B cell (illustrated as CD19, gray) presents both ligand and receptor molecules. Prognosis is determined by the directionality of the signal upon binding of the complementary molecule, which is expressed by other cells in the surroundings. (B) Proportion of dual interactions established between each possible immune pair under study and their distribution in each prognostic condition. (C) Heat map showing dual interactions with potential targetability in MM. Displayed are the percentage of cells expressing the molecule and the median expression of both molecules by the pair of engaged cell types. Highlighted are those interactions which are significant (P < .05) in 1 condition but not the other. In the shown interactions, the first molecule is expressed in all cases by CD19+ cells (B cells); the second molecule is expressed by the 4 cell types (CD3+, CD14+, CD19+, or CD45+TN). On the left side of the list of interactions, the column represents TTPlow patients; on the right side, the column represents TTPhigh patients. (D) Model illustrating IL-15–IL-15RA interactions on diverse cell types as example.
To identify candidate interactions with therapeutic potential, we focused on molecules expressed at the cell surface. We hypothesized that these molecules might represent potential therapeutic targets, being accessible at the cell surface and potentially targetable by immunotherapy/chimeric antigen receptor (CAR) T cells. We identified a subset of 13 surface-restricted dual interactions that may serve as potential therapeutic targets (Figure 4C). For instance, we identified an interaction of interleukin-15 (IL-15) and IL-15RA among the dual interactions. When IL-15 is expressed by the B cell and interacts with IL-15RA on the CD3+ or CD45+TN cells, it confers a favorable prognosis. Transpresented IL-15 sensed by NK cells and T cells triggers immune activation and cytotoxicity.39-42 However, in patients with short TTP, both the receptor IL-15RA and the ligand IL-15 are expressed by CD19+ cells and CD14+ cells, suggesting the possibility of bidirectional signaling (Figure 4C). IL-15 transpresentation involving CD14+ cells has been reported to induce the secretion of proinflammatory cytokines and chemoattractants, such as IL-1, IL-6, IL-8, TNF-α, and MCP-1 (Monocyte Chemoattractant Protein-1).42-44 Therefore, the same interaction can result in increased antitumor immunity or in the creation of a proinflammatory immunosuppressive microenvironment, depending on the cell types involved. We further validated the expression of these molecules in the relevant interacting cell types using the Human Protein Atlas and Bloodspot (Blueprint data set; supplemental Figure 4A-B). These findings suggest a model in which targeting a specific receptor on the cell surface may not be beneficial for patients. Instead, a distinct cell type expressing this molecule should be targeted (Figure 4D). This can be achieved using various novel strategies in immunotherapy, such as the SynNotch system,45 which would allow for the targeting of a molecule on a particular cell type.
Characterizing interacting cell types with single-cell resolution
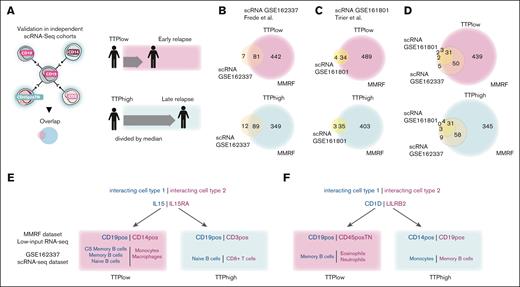
To validate these interactions, we used 2 independent published scRNA-seq data sets,27,28 which interrogated the bone marrow immune microenvironment in patients with R/R MM receiving IMiD-based treatments. We performed cell interaction analyses in these cohorts and assessed the overlap with our data set (Figure 5A). We identified a lower number of interactions in the scRNA-seq data sets (supplemental Figure 5A) than in our low-input RNA-seq analyses, presumably related to the fact that a greater number of genes are detected in minipools than in single cells. Despite this and the fact that the validation data sets were from patients with R/R MM rather than ND patients, the majority of predicted interactions identified overlapped with those derived from the low-input RNA-seq (Figure 5B-C). Most of the interactions were detected in all 3 data sets independently of interacting cell types (Figure 5D). We further included an additional data set of patients with NDMM who went on to receive IMiD treatment29 and found that the majority of predicted interactions identified overlapped with those derived from the low-input RNA-seq (supplemental Figure 5B).
Validation of prognostically relevant cell-cell interactions in independent single-cell sequencing cohorts. (A) Diagram. (B-D) Venn diagrams showing overlap between interactions identified in TTPlow/TTPhigh patients of our cohort with published cohorts using single-cell RNA-seq. (E-F) Examples of dual interactions that were detected with statistically significance in our cohort and published cohorts using single-cell RNA-seq. CS Memory B cells, class-switched memory B cells.
Validation of prognostically relevant cell-cell interactions in independent single-cell sequencing cohorts. (A) Diagram. (B-D) Venn diagrams showing overlap between interactions identified in TTPlow/TTPhigh patients of our cohort with published cohorts using single-cell RNA-seq. (E-F) Examples of dual interactions that were detected with statistically significance in our cohort and published cohorts using single-cell RNA-seq. CS Memory B cells, class-switched memory B cells.
To investigate whether these interactions are IMiD specific, we next investigated 2 data sets from patients who received non-IMiD–based treatments: GSE21007930 for a BCMA (B-Cell Maturation Antigen)–CAR T-cell–treated patient population; and GSE16180128 consisting of patients who received either MEKi (Mitogen-Activated Protein Kinase Kinase inhibitor) or proteasome inhibitor treatment. We found that most of the detected interactions from the non-IMiD–based treatments overlap with our low-input RNA-seq data set, arguing that most interactions are not treatment specific (supplemental Figure 5C-F). Although most interactions were shared between data sets, regardless of treatment, this was not the case for dual interactions. In fact, the dual interactions predicted from our low-input RNA-seq data set were not found as dual interactions in any of the other inspected data sets (IMiD or non-IMiD treated), arguing that these are specific to the data set and the context (supplemental Figure 5G). However, discrepancies in the predicted number of interactions could affect the recovery of identical dual interactions.
To further delineate the immune compartments and pinpoint the specific cell types involved in interactions linked to favorable and unfavorable outcomes at the single-cell level, we leveraged a published single-cell data set (GSE162337) profiling myeloma and immune cells from 9 patients with R/R MM receiving Elo-PVD (Elotuzumab, pomalidomide, bortezomib and dexamethasone) treatment.27 Two interactions that were identified as dual interactions in both data sets are CD1D/LILRB2 and IL-15/IL-15RA. The interaction of IL-15/IL-15RA plays a role in adaptive and innate immunity and has been shown to be involved in myeloma progression.42,46 Notably, in our low-input RNA-seq data set, TTPhigh patients showed enriched IL-15/IL-15RA interaction, specifically enriched between CD19+ cells interacting with CD3+ or CD45+TN cells, whereas in TTPlow patients, the interaction was identified between CD19+ cells or between CD19+ and CD14+ cells (Figure 4C). Similarly, in the scRNA-seq–derived data set,27 we identified the IL-15/IL-15RA interaction between CD19+ and CD3+ cells in the TTPhigh condition and were able to further define these cell types as naive B cells and CD8+ T cells. In the TTPlow patients, this same interaction was also found between CD19+ and CD14+ cells, and we defined these as (class-switched) memory B cells or naive B cells and monocytes or macrophages, respectively (Figure 5E).
Regarding CD1D/LILRB2, we detected an interaction between CD14+ and CD19+ cells in TTPhigh patients and identified the interacting cell types as monocytes and memory B cells in the scRNA-seq data sets. However, in TTPlow patients, we recovered the same interaction between CD19+ and CD45+TN cells and identified memory B cells and eosinophils/neutrophils as the specific cell types driving this interaction (Figure 5F). Therefore, by interrogating a distinct data set with single-cell resolution, we were able to validate the “dual” effect of candidate pathways, depending on the specific cell types involved. Lastly, to determine whether RNA expression could serve as a proxy for protein expression, we evaluated how RNA expression corresponded to protein expression in single cells measured by CITE-seq for 49 different markers in a data set assessing bone marrow myeloma and immune cells after BCMA CAR T-cell therapy.30 We found that RNA levels generally reflected protein expression accurately (supplemental Figure 6A-B), particularly for molecules well established to define hematopoietic lineage and differentiation (supplemental Figure 6C-F).
Discussion
Our study aimed to explore whether activating the same immune signaling pathway can yield different results. Our data imply that although activation of a pathway through interaction with 1 cell type can yield positive effects, its activation through interaction with another cell type can lead to negative effects.
We examined the immune microenvironments of patients with NDMM who relapsed after IMiD-based therapy, because there is ample evidence for the interaction and the interdependence of MM cells and the immune microenvironment.28,47-51 Using FACS, low-input RNA-seq, and computational deconvolution, we detected a shift from naive B cells to more differentiated memory B cells in patients with a worse prognosis, similar to changes we observed when comparing patients with myeloma to normal donors. Our pathway enrichment analysis revealed that B-cell differentiation–promoting interleukin signals, such as IL-7, IL-10, and IL-15, and downstream JAK/STAT signaling were enriched in the B-cell compartment of patients with poor prognosis.52-56 Our study therefore highlights the crucial role of the B-cell compartment in response to IMiD-based therapy in patients with NDMM. These findings and the evolving understanding of B-cell contributions to antitumor immunity were the reasons why we focused on B-cell compartment in this study.
In myeloma, long-term immunosurveillance is critical. Many immunotherapies stop working, although the antigen is still expressed by the target cells, pointing to a dysfunctional immune response. Growing evidence indicates that T-cell exhaustion due to prolonged antigen exposure results in an immune system unable to efficiently combat tumors.57,58 Lower tumor burdens and less immune activation may help maintain a more competent immune system, leading to a better prognosis. Our findings suggest that patients with good prognosis maintain a less differentiated B-cell compartment, with naive B cells and cytotoxic T and NK cells, whereas those with a poor prognosis exhibit a more differentiated B-cell compartment that may promote a proinflammatory microenvironment.
Leveraging computational inference, we uncovered a complex network of cell-cell interactions within the immune system, pinpointing those that correlate with prognosis. Interestingly, some of these interactions are found in opposite prognostic conditions, conferring either favorable or poor prognosis depending on the specific cell types involved. Such signal ambiguity, as is known for IL-2 signaling,59 which can promote T effector cells and immunosuppressive Tregs at the same time, complicates these signals' characterization and targeting. Thus, understanding the cell types involved in these interactions can provide clearer insight into resistance mechanisms. For instance, we identified a dual role for the interaction of IL-15/IL-15RA. When IL-15 expressed by B cells interacts with IL-15RA on T or NK cells, it fosters a favorable prognosis, enhancing immune activation and cytotoxicity, and boosting CD8+ memory T-cell efficacy.39-42 Conversely, IL-15 signaling involving CD14+ cells can promote proinflammatory cytokines and chemoattractants, such as IL-1, IL-6, IL-8, TNF-α, and MCP-1,42-44 creating a proinflammatory immunosuppressive microenvironment. Thus, the outcome of the same interaction may vary based on the participating cell types.
These observations underscore the importance of considering specific cell types when targeting ligand-receptor interactions. Understanding this concept is of paramount importance for the development of more selective immunotherapies. Our findings advocate for a paradigm shift toward selectively targeting cells expressing specific molecules. Although traditional immunotherapy indiscriminately targets surface receptors, emerging technologies, such as CAR T cells, permit cell type–specific targeting of surface receptors.45 This capability of conditional cellular therapies facilitates a more precise manipulation of the immune microenvironment and a promising avenue for optimizing therapeutic interventions, which allows for the specific targeting of detrimental immune signaling pathways while preserving essential immune functions and mitigating toxicities.
Given that cancer immunotherapies yield lasting responses in only a fraction of patients, there is a need for devising novel immunotherapeutic strategies. Although T cells and NK cells have so far been the primary focus of immunotherapeutic interventions, recently it has been suggested that the manipulation of TIM-1 (T cell immunoglobulin and mucin domain-1)–expressing B cells may facilitate the activation of the second arm of adaptive immunity, leading to improved antitumor immune responses.60 Leveraging the diverse functions of B cells, including antigen presentation and production of antibodies, may open avenues for novel immunotherapeutic approaches, including combination therapies that exploit both T-cell and B-cell responses. Therefore, investigating the potential role of B cells is crucial for developing more comprehensive immunotherapeutic strategies. Further exploration of the interplay between B cells and other immune components within the tumor microenvironment therefore holds great promise for advancing the field of cancer immunotherapy.
In summary, our research demonstrates how diverse immune microenvironments in patients with NDMM influence their responses to IMiDs and other therapies. We uncover the intricate signaling network within the bone marrow immune microenvironment, which critically affects diverse immune cell functions. We further explain how different outcomes can result from the same molecular interaction, based on the participating cellular identities. Moving forward, selectively targeting cells expressing certain molecules, rather than just targeting the molecules themselves, is crucial to balance beneficial and harmful signals, ensuring effective long-term tumor immunity.
Acknowledgments
Funding for this research was provided by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co, Inc, Rahway, NJ, and the Multiple Myeloma Research Foundation.
Authorship
Contribution: P.H-L., J.F., P.A., and T.V. designed and performed data analysis; D.A. provided samples and advice; J.F., B.K., and J.G.L. designed experiments, provided project leadership, supervised the analysis, and wrote the manuscript; and all authors discussed the results and reviewed the manuscript.
Conflict-of-interest disclosure: J.G.L. acknowledges the support of research funding for an unrelated project from Bristol Myers Squibb and the receipt of honoraria for consulting services provided to Asher Bio. The remaining authors declare no conflicting financial interests.
Correspondence: Julia Frede, Department of Medical Oncology, Jerome Lipper Multiple Myeloma Center, Dana-Farber Cancer Institute, Boston, MA 02215; email: julia_frede@dfci.harvard.edu; Jens G. Lohr, Department of Medical Oncology, Jerome Lipper Multiple Myeloma Center, Dana-Farber Cancer Institute, Boston, MA 02215; email: jensg_lohr@dfci.harvard.edu; and Birgit Knoechel, Department of Pediatric Oncology, Dana-Farber Cancer Institute, Boston, MA 02215; email: birgit_knoechel@dfci.harvard.edu.
References
Author notes
J.F., B.K., and J.G.L. jointly supervised this work.
The sequencing data used in this study have been deposited in the Gene Expression Omnibus database (accession number GSE263702).
The full-text version of this article contains a data supplement.