Key Points
Integrated clonotype and mRNA expression levels in 6 patients, identifying common and divergent transcriptional states.
Characterization of transcriptional changes with progression on photopheresis and vorinostat.
Abstract
Cutaneous T-cell lymphomas (CTCLs) are a spectrum of diseases with varied clinical courses caused by malignant clonal proliferation of skin-tropic T cells. Most patients have an indolent disease course managed with skin-directed therapies. In contrast, others, especially in advanced stages of disease or with specific forms, have aggressive progression and poor median survival. Sézary syndrome (SS), a leukemic variant of CTCL, lacks highly consistent phenotypic and genetic markers that may be leveraged to prevent the delay in diagnosis experienced by most patients with CTCL and could be useful for optimal treatment selection. Using single-cell mRNA and T-cell receptor sequencing of peripheral blood immune cells in SS, we extensively mapped the transcriptomic variations of nearly 50 000 T cells of both malignant and nonmalignant origins. We identified potential diverging SS cell populations, including quiescent and proliferative populations shared across multiple patients. In particular, the expression of AIRE was the most highly upregulated gene in our analysis, and AIRE protein expression could be observed over a variety of CTCLs. Furthermore, within a single patient, we were able to characterize differences in cell populations by comparing malignant T cells over the course of treatment with histone deacetylase inhibition and photopheresis. New cellular clusters after progression of the therapy notably exhibited increased expression of the transcriptional factor FOXP3, a master regulator of regulatory T-cell function, raising the potential implication of an evolving mechanism of immune evasion.
Introduction
Cutaneous T-cell lymphomas (CTCLs) are a group of heterogeneous malignancies of T-cell origin that traffic to the skin. Mycosis fungoides (MFs) and Sézary syndrome (SS) are CTCLs comprised of skin-tropic CD4+ T cells with varying degrees of blood involvement.1,2 Despite the clonal nature of both MF and SS, the course of the diseases can be highly variable, with a subset possessing a relatively indolent disease course, whereas other patients manifest widespread disease burden beyond the skin; in particular, patients with SS exhibit prominent blood involvement and carry a poor prognosis. Advanced stages of CTCL have a 5-year survival as low as 24%,1 with patients often cycling through therapies that do not offer long-term durable responses. Thus, a deeper understanding of the CTCL disease process and determinants of tumor behavior are critical unmet knowledge gaps that could assist in determining optimal therapeutic regimens for patients with SS and MF.
Although CTCL represents a clonal proliferation of malignant T cells, we and others have previously demonstrated single-cell transcriptional heterogeneity of the circulating malignant T-cell population of patients with SS.3,4 This heterogeneity has also been reported from single-cell sequencing of CTCL skin lesions, demonstrating marked intertumoral and intratumoral transcriptional heterogeneity.5 One theory for the differential presentation and course of CTCL for patients is the varied composition of distinct subsets of malignant T cells.2 Single-cell sequencing enables the investigation of distinct subset analysis for the leukemic SS, which may shed light on niche development and potential vulnerabilities to specific treatments.
The transcriptional heterogeneity of CTCL may explain the extensive treadmilling of therapeutic response and recurrence. Histone deacetylase inhibitors (HDACi) are a mainstay of CTCL treatment and are thought to alter key genetic programs involved in tumor development and progression.6 However, the trials for which the approval of HDACi was based found only 30% to 35% of patients with CTCL respond to HDACi, with a median duration of response of 13.7-15 months.7,8 Interestingly, an in-depth analysis of HDACi in patients with SS found highly individualized responses in not only T-cell number but variable genomic and mRNA expression changes.9,10 Brentuximab vedotin (BV), another therapy for CTCL, consists of a chimeric antibody directed against CD30, a cell marker variably found in MF/SS, conjugated to monomethyl auristatin E, an inhibitor of microtubule polymerization and hydrolysis. Studies examining the efficacy of BV in advanced-stage CTCL have indicated that there is no correlation between tumoral CD30 expression and time to response, duration of response, progression-free survival, and event-free survival11,12 supporting that other patients or tumor-specific features play a role. These data underscore the need for a greater understanding of the maintenance of the malignant potential of CTCL and the need for better predictive biomarkers in patients with CTCL undergoing treatment.
We used single-cell mRNA and T-cell receptor (TCR) sequencing across isolated CD45+ immune cells in the peripheral blood of 6 patients with SS. With over 50 000 single cells, our studies revealed extensive common and differential gene expression markers and cellular pathway alterations within malignant SS T cells. In addition to this analysis, our study uncovered alterations at the single-cell level of a patient with SS during treatment with HDACi and photopheresis through therapeutic failure and disease progression, offering a novel insight into resistance mechanisms for combinatorial therapy. Therefore, these data indicate that determinations of tumor heterogeneity and composition by transcriptionally distinct malignant cell subpopulations may hold predictive value in treatment response and utility in clinical decision-making and therapeutic choice.
Methods
Patient recruitment
The current study was approved by the University of Iowa and the Mayo Clinic Institutional Review Board and conducted under the Declaration of Helsinki Principles. The patients were recruited from the Department of Dermatology Cutaneous Lymphoma Clinic at the University of Iowa and the Cutaneous Lymphoma Clinic at the Mayo Clinic in Scottsdale, Arizona. Samples from the Mayo Clinic were shipped overnight at 4°C to the University of Iowa. Informed written consent was received from the participant before inclusion in the study.
Flow cytometry
Blood draws were performed, and peripheral blood mononuclear cells were isolated using a Ficoll gradient. Cells were labeled for CD45 and flow-sorted to isolate immune cells on a Becton Dickinson Aria II. The Patient 1 sample isolation involved flow-sorted peripheral blood CD4+ T cells only, and the baseline data were previously described.3
Single-cell RNA sequencing
Sequencing for 5′ gene expression and T-cell receptor was performed using the Chromium (10x Genomics, Pleasanton, CA) and Illumina (San Diego, CA) sequencing technologies. Amplified cDNA was used to construct both 5′ expression and TCR enrichment libraries. Libraries were pooled and run on separate lanes on an Illumina HiSeq 4000. Each lane consisted of 150 bp paired-end reads. Basecalls were converted into FASTQs using the Illumina bcl2fastq. FASTQ files were aligned to the human genome (GRCh38) using the CellRanger v3.0.1 pipeline as described by the manufacturer. The TCR V(D)J sequences were aligned to the vdj_GRCh38_alts_ensembl genome build provided by the manufacturer.
Single-cell data processing and analysis
Initial processing of immune cells from peripheral blood of 6 patients used the Seurat R package (v3.0.2). Single cells used in subsequent analysis were comprised of CTCL1 (n = 7828), CTCL2 (n = 32 592), CTCL3 (n = 2250), CTCL4 (n = 1479), CTCL5 (n = 1090), and CTCL6 (n = 2677). CTCL1 sequencing data were derived from the previous publication3 and included as part of the baseline/integrated data. For this sample, we sequenced a second time point after progression on vorinostat (n = 7127) which has not been previously published. Samples were combined into a single Seurat object using canonical correlational analysis and mutual nearest neighbors.13,14 Dimensional reduction to form the Uniform Manifold Approximation and Projection (UMAP) plots used the top 40 calculated dimensions and a resolution of 0.5. Cluster markers and differential gene expression analyses were performed using the Wilcoxon rank-sum test with an unsupervised approach involving no gene filtering. FOXP3+ cells were defined as individual cells with FOXP3 counts ≥1. Percent expression graphs were created using the schex R package (v1.1.5, development version), setting nbins = 80 and in the make_hexbin() call. Cell trajectories were created using the monocle (v2.10.1) R package15 using the top 4750 genes for ordering and the DDRtree dimensional reduction strategy. In addition, the individual time points were used as the residual model string during the reduction of dimensions to eliminate differences based on the batch effect. Cell cycle scoring was performed on single cells using the previously described gene set16 and the CellCycleScoring function in the Seurat package.
Single-cell immune phenotyping used the SingleR (v1.0.1) R package17; the cell-type based correlations and single-cell expression values were compared with transcriptional profiles from pure cell populations in Human Primary Cell Atlas.18 Single-sample gene set enrichment analysis (ssGSEA) used the escape R package.19 Individual gene sets were derived from the GSEA Hallmark library,20 Kyoto Encyclopedia of Genes and Genome,21 and Biocarta. Expression data were visualized using the ggplot2 (v3.2.1) and pheatmap (v1.0.12) R packages. TCR sequencing results were processed using the scRepertoire (v1.0.0).22 Malignant clones were defined as clones with immune repertoire occupancy >0.1. Genes upregulated at time point 2 (n = 1428) underwent GSEA using the Enrichr software23 using the same libraries as above. Results with adjusted P value < .25 are presented in supplemental Table 6. Processed data are available in the GEO accession: GSE124899 and GSE146586.
Bulk-sequencing analysis
Raw expression data for GSE11311324 were downloaded from the National Center for Biotechnology Information Sequence Read Archive and converted to FASTQ files using the SRA toolkit. Samples were aligned with the kallisto pseudoalignment25 and GRCh38 build for the human genome, aggregating the estimated counts for transcripts into gene-level quantifications based on Human Genome Organization Gene Nomenclature Committee gene symbol. Sample expression values were processed using the Sleuth R Package (v0.30.0). ssGSEA for the bulk samples was performed using the GSVA (v1.30.0) R package using the Poisson function for cumulative distribution. Immune receptor quantification for bulk RNA sequencing was performed using MiXCR using the default pipeline.26 Recovered immune receptor reads were then filtered for the dominant T-cell receptor V gene, corresponding to either ɑβ or γδ T cells, and normalized by the entire recovered immune receptor repertoire.
Immunohistochemistry
Immunohistochemistry was performed on a Dako Autostainer Link 48 after deparaffinization, rehydration, and heat-induced epitope retrieval on a Dako PT Link. All antibodies were retrieved in Dako Target Retrieval Solution, HpH (EDTA-based). All primary antibody incubations were of a 30-minute duration. The polymer-based Dako EnVision FLEX kit was used for detection; all detections were 30 minutes. Diaminobenzidine was used as the chromogen. Slides were subsequently lightly counterstained with Harris hematoxylin, dehydrated, and coverslipped for pathologist review. The primary antibodies used in this study included CD3 (Dako; polyclonal; 1:800), CD4 (Novocastra; clone 4B12; 1:100), and autoimmune regulator (AIRE; Invitrogen; clone 5H12; 1:50). A multitissue block, including tonsil, was used as the positive tissue control for CD3 and CD4, while thymus was used for AIRE. Nuclear AIRE staining was quantified as mean positive cells across a minimum of 5 high-power fields.
Statistical analysis
Statistical analyses were performed in R (v3.6.1). Two-sample significance testing used Welch’s t test, with significance testing for >3 samples using one-way analysis of variance (ANOVA) with Tukey honest significance determination for correcting multiple comparisons. Unless otherwise noted in the figure legend, significance for gene expression is based on the cutoff of log-fold change (LFC) ≥1 or ≤−1 and an adjusted P value < .05. Single-cell relative percentages by categorical variables used the total number of cells in the indicated sequencing run as a denominator for normalization. Analysis of the distribution of cells across multiple categorical variables used the χ2 test.
Results
Sequencing results of the peripheral blood of 6 patients with SS
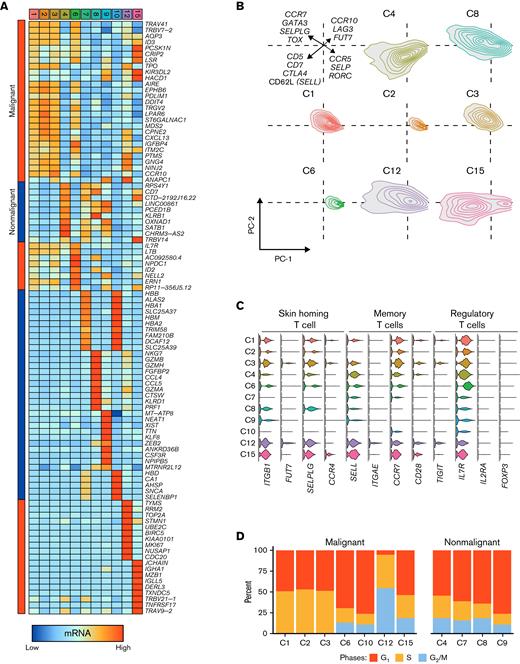
A total of 54 994 cells were sequenced and passed, filtering from the peripheral blood of 6 separate patients with SS (Figure 1A). Patient information and a summary of the sequencing results are available in supplemental Table 1. Based on mRNA expression, we observed 16 distinct clusters. Using both the percent of cells expressing lineage markers (Figure 1B) and correlations (Figure 1C) based on the human primary cell atlas, we identified 11 T-cell clusters (C1, C2, C3, C6, C7, C8, C9, C10, C12, and C15), 1 B-cell cluster (13C), and 3 myeloid-cell clusters (C5, 11C, and C14). The majority of the sequenced peripheral cells were T cells, with a mean percent of T cells per patient equal to 76.9 ± 15.2% (Figure 1D). Myeloid and B cells comprised a mean percentage of 19.2 ± 12.7% and 3.9 ± 3.9%, respectively. Each patient sample varied in the distribution of cell types (Figure 1E). Notably, the Patient 2 sample consisted of a relatively high number of T cells in the C2, C3, and C6 T-cell clusters (Figure 1E), while Patient 5 and 6 samples had a wide distribution of cells across all 3 cell types (Figure 1E).
Single-cell map of peripheral blood across 6 patients with Sézary syndrome. (A) UMAP projection of the flow-sorted CD45+ immune cells (n = 54 994) across 6 patients: T cells/CTCL cells (Cluster 1, 2, 3, 4, 6, 7, 8, 9, 10, 12, and 15), myeloid cells (Cluster 5, 11, and 14), and B cells (Cluster 13). (B) Percent of cells expressing selected lineage markers for T cells (CD2 and CD3G), myeloid cells (ITGAM [CD11b] and ITGAX [CD11c]), and B cells (CD19 and CD79A). (C) Normalized correlation values across rows ranging from 0 to 1 for predicted immune cell phenotypes from the Human Primary Cell Atlas18 based on the SingleR R package17 for each cluster. (D) Percentage of total cells for each patient by cell type. (E) Cell distribution by patient sample both as contour graphs and summarized in the accompanying bar chart by patient and cluster. Clusters representing >15% of total patient cells are labeled.
Single-cell map of peripheral blood across 6 patients with Sézary syndrome. (A) UMAP projection of the flow-sorted CD45+ immune cells (n = 54 994) across 6 patients: T cells/CTCL cells (Cluster 1, 2, 3, 4, 6, 7, 8, 9, 10, 12, and 15), myeloid cells (Cluster 5, 11, and 14), and B cells (Cluster 13). (B) Percent of cells expressing selected lineage markers for T cells (CD2 and CD3G), myeloid cells (ITGAM [CD11b] and ITGAX [CD11c]), and B cells (CD19 and CD79A). (C) Normalized correlation values across rows ranging from 0 to 1 for predicted immune cell phenotypes from the Human Primary Cell Atlas18 based on the SingleR R package17 for each cluster. (D) Percentage of total cells for each patient by cell type. (E) Cell distribution by patient sample both as contour graphs and summarized in the accompanying bar chart by patient and cluster. Clusters representing >15% of total patient cells are labeled.
Defining malignant T cells in the peripheral blood of patients with SS
Across the 11 T-cell clusters (Figure 2A), we next explored the expression patterns of common T-cell and CTCL markers (Figure 2B). Although all clusters expressed CD3D and CD5, pan-T–cell genes, there was delineation in the expression of CD4 (high clusters: C1, C2, C3, and C6) and CD8A (high clusters C8; moderate clusters C4, C7, C9, C10). A general absence of CD7, a common feature in CTCL, was observed in clusters C1-3 and C6 (Figure 2B). These clusters possessed prominent expression of CCR4, CXCL13, and PD-1 (PDCD1) (Figure 2B), genes previously associated with SS.3,5,27,28 Accompanying the mRNA classification of malignant and nonmalignant T cells, we also used the V(D)J single-cell sequencing results for the TCR, recovering the sequences for 43 074 T cells (89.9%). Using the scRepertoire22 R package, we defined clonotypes as both the sequence of genes and the nucleotide sequence of both the TCR A and B chains (supplemental Figure 1A); a variety of unique clonotypes were isolated across the 6 samples (supplemental Figure 1B). The number of unique clonotypes sequenced did not predict the clonal homeostasis or occupied repertoire space for each sample, with 95.2% of Patient 2 T cells formed from the dominant clonotype for that sample despite having 1254 unique clonotypes (supplemental Figure 1C,D).
Differentiating malignant and nonmalignant T cells. (A) UMAP projection of CD3+ T-cell clusters. (B) Percent of cells expressing selected common markers for CTCL across the UMAP projection. (C) UMAP projection with proportion of total reads per sample of TCR V(D)J-based clonotype attached into the following grouping: hyperexpanded (0.1 < X ≤ 1), large (0.01 < X ≤ 0.1), medium (0.001 < X ≤ 0.01), small (1e−4 < X ≤ 0.001), and rare copies (0 < X ≤ 1e−4) using the scRepertoire R package22 with the percentage of cells in each clonotype category in the lower bar chart. Malignant T-cell clusters (C1, C2, C3, C6, C10, C12, and C15) had >50% of cells with hyperexpanded clonotypes. (D) Percent difference (Δ percent) between malignant and nonmalignant T cells vs log2-fold change with the top 10 significant (Bonferroni P value < .05) upregulated genes by both percentages and fold change labeled. (E) Violin plot of relative mRNA expression of the top 12 significant genes by Δ percent difference between malignant (M) and nonmalignant (NM) T cells. (F) Significantly upregulated genes in the single-cell (SC) cohort and a secondary SS cohort29 with the top 10 genes by a log-fold change in overlapping comparison and unique to the SC cohort. (G) Representative immunohistochemistry images for SS at 400× magnification. Representative images and data summary (bar graph) of 35 patients with MF, SS, or T-cell NHL. Black arrows indicate nuclear staining of AIRE, and the images displayed are at 200× magnification.
Differentiating malignant and nonmalignant T cells. (A) UMAP projection of CD3+ T-cell clusters. (B) Percent of cells expressing selected common markers for CTCL across the UMAP projection. (C) UMAP projection with proportion of total reads per sample of TCR V(D)J-based clonotype attached into the following grouping: hyperexpanded (0.1 < X ≤ 1), large (0.01 < X ≤ 0.1), medium (0.001 < X ≤ 0.01), small (1e−4 < X ≤ 0.001), and rare copies (0 < X ≤ 1e−4) using the scRepertoire R package22 with the percentage of cells in each clonotype category in the lower bar chart. Malignant T-cell clusters (C1, C2, C3, C6, C10, C12, and C15) had >50% of cells with hyperexpanded clonotypes. (D) Percent difference (Δ percent) between malignant and nonmalignant T cells vs log2-fold change with the top 10 significant (Bonferroni P value < .05) upregulated genes by both percentages and fold change labeled. (E) Violin plot of relative mRNA expression of the top 12 significant genes by Δ percent difference between malignant (M) and nonmalignant (NM) T cells. (F) Significantly upregulated genes in the single-cell (SC) cohort and a secondary SS cohort29 with the top 10 genes by a log-fold change in overlapping comparison and unique to the SC cohort. (G) Representative immunohistochemistry images for SS at 400× magnification. Representative images and data summary (bar graph) of 35 patients with MF, SS, or T-cell NHL. Black arrows indicate nuclear staining of AIRE, and the images displayed are at 200× magnification.
T-cell clonal frequency was then placed into categories by copies within the patient sample. TCR clonal categories were organized by proportion of total productive TCR reads per patient with hyperexpanded (0.1 < X ≤ 1), large (0.01 < X ≤ 0.1), medium (0.001 < X ≤ 0.01), small (1e−4 < X ≤ 0.001) and rare clonotypes (0 < X ≤ 1e−4). The C1, C2, C3, C10, C12, and C15 clusters possessed T cells with >90% hyperexpanded clonotypes, so-called malignant clusters (Figure 2C), with each sample having a single, dominant clone. Notably, nonmalignant clusters were C4, C7, C8, and C9. To investigate potential novel markers or therapeutic targets of the peripheral SS T cells, we next performed differential gene expression analysis across T cells with a hyperexpanded clonotype compared with T cells with single, small, medium, or large clonotypes. Unlike bulk sequencing approaches, SC mRNA sequencing allows for the comparison of differential genes in the context of the percent of cells expressing the gene or genes of interest. This allows for not only the difference in terms of LFC but also in terms of the difference in the percent of malignant vs nonmalignant T cells expressing the gene(s) (Δ percent). Non-TCR genes with the highest LFC and greatest discrimination between malignant vs nonmalignant T cells were AIRE (LFC = 2.42; Δ = 49.2%), NEDD4L (LFC = 2.39; Δ = 28%), IGFBP4 (LFC = 2.28; Δ = 22.4%), TUSC3 (LFC = 2.24; Δ = 16.7%), and PDLIM1 (LFC = 2.20; Δ 28.1%) (Figure 2D,E). Conversely, CD2 (LFC = −0.66; Δ = 35.2%), CD7 (LFC = −2.21; Δ = 58.3%), and CD49f (ITGA4, LFC = 3.14; Δ = 30.3%) were among the most downregulated genes in the malignant SS T cells (supplemental Figure 2). The complete results for the differential expression analysis are available in supplemental Table 2.
We also compared our list of differentially upregulated genes (n = 284) to previously identified upregulated genes in peripheral blood of SS (n = 53).29 The comparison yielded an overlap coefficient of 49.1% and 26 common genes shared between the analyses (Figure 2F). Using a 50-patient bulk RNA cohort of normal skin, MF, and SS biopsies,24 we first established the relationship of clonality and diversity in the samples (supplemental Figure 3A,B). In this cohort, 237 of the 284 genes were expressed, and we found 72 of the 237 genes significantly and directly correlated with the proportion of dominant V-gene recovered, a marker of clonality (supplemental Figure 3C). Of the genes with nonsignificant increased expression in the single-cell cohort and expressed in the bulk sequencing data (n = 2594), 532 genes were positively correlated with dominant V-gene proportion. Contingency comparison by Fischer’s exact test showed a strong association between the single-cell differentially-expressed genes and correlations (P = 6.47e−4). We also found that 48 of the 284 genes inversely correlated with the diversity of the immune repertoire (supplemental Figure 3C). Top genes, ranked by LFC in both comparisons (Figure 2F, left list) and by LFC in our cohort (Figure 2F, right list), are displayed. Half of these top 20 genes were significantly correlated with clonality (directly) or diversity (inversely) in the secondary bulk RNA cohort (supplemental Figure 3D,E). We chose to survey skin biopsies of MF (n = 17), SS (n = 5), and T-cell non-Hodgkin’s lymphoma (NHL; n = 13) patients using IHC for one of these novel markers AIRE (Figure 2G). Nuclear staining of AIRE ranged across the patients, and we found AIRE in 11 of 17 MF and 5 of 6 SS samples (Figure 2H). Although the mean density of AIRE+ SS was varied, a comparison between CTCL staining (MF + SS) and NHL produced a P value of .005321.
Transcriptional heterogeneity within malignant and nonmalignant T cells
We next wanted to understand the differences in the gene expression that produced the different T-cell clusters. Although the malignant clusters C1, C2, and C3 shared a number of common gene markers described above (Figure 3A), other malignant clusters expressed unique markers. For example, C12 had high levels of expression of TOP2A and MKI67, markers of cell-cycle activation, compared with any other T-cell cluster (Figure 3A). The nonmalignant cluster, C8, had high granzyme and perforin expression levels, consistent with cytotoxic functions (Figure 3A) and its high expression of CD8A. To better understand the possible implications of the differential gene expression across the T-cell clusters, we performed principal component analysis (PCA) on scaled mRNA levels of genes involved in T-cell development and differentiation (Figure 3B), with the full list of genes available in supplemental Table 3. In the malignant C1, C3, and C12 clusters, the PCA-identified expression consisted of GATA3, CCR7, and TOX expression (Figure 3B). The nonmalignant C8 cluster was also unique in the high expression of cytotoxic markers, such as GZMA, GZMB, and PRF1, while detectable, albeit low levels of expression of these mRNA species were only seen in the malignant C12 cluster (Figure 3A). We also examined selected canonical gene markers of CTCL transcriptional profiles. Among the skin trafficking markers, C12 and C15 had the highest expression of SELPLG (CLA precursor) and the fucosyl-transferase, FUT7,30 although they possessed lower chemokine receptor levels, CCR4 (Figure 3C). Memory phenotypes have also previously been described in CTCL31,32; high expression of CCR7 across malignant T cells compared with nonmalignant T cells is consistent with prior reports of SS cells either emerging from, or at the least possessing, central memory T–cell-like characteristics.33 Although we have previously described both clonal central memory and FOXP3+ regulatory T-cell (Treg) clusters examining a single patient with SS,3 the aggregated single-cell analysis failed to reveal a representative cluster for Treg or Treg-like cells (Figure 3C), possibly due to the increased number of cells, the inclusion of additional immune cell types, or drop-out effect of single-cell sequencing. With the exception of CTCL3 and CTCL6 samples, FOXP3+ T cells were clustered across C1, C3, and C4, forming a range of 2.7% to 38.9% of the cells in the clusters by patient ID.
Heterogeneity in both normal and malignant SS T cells. (A) Heatmap of the top 10 genes for each of the single-cell clusters with Bonferroni P value < .05. Bars on the left indicate malignant (red) or nonmalignant (blue) predominance. (B) Density plots of the principal component analysis (PC1 and PC2) of genes involved in T-cell differentiation. (C) Selection of genes by cluster identity for skin-homing, central memory, and Treg phenotypes (IL7R-low/negative associated with Tregs). (D) Percent of cells in each cluster in phases of the cell cycle by regression analysis. (E) ssGSEA enrichment scores for gene signatures derived from the malignant SS clusters applied to a mixed MF/SS bulk RNA cohort.24 (F) The proportion of dominant V read vs ssGSEA enrichment scores for gene signatures derived from the malignant SS clusters applied to a mixed MF/SS bulk RNA cohort.24 (G) Z-score scaled ssGSEA enrichment scores for Biocarta, Kegg, and Hallmark libraries by single-cell cluster. The median values across nonmalignant (NM) clusters are reported in a single column.
Heterogeneity in both normal and malignant SS T cells. (A) Heatmap of the top 10 genes for each of the single-cell clusters with Bonferroni P value < .05. Bars on the left indicate malignant (red) or nonmalignant (blue) predominance. (B) Density plots of the principal component analysis (PC1 and PC2) of genes involved in T-cell differentiation. (C) Selection of genes by cluster identity for skin-homing, central memory, and Treg phenotypes (IL7R-low/negative associated with Tregs). (D) Percent of cells in each cluster in phases of the cell cycle by regression analysis. (E) ssGSEA enrichment scores for gene signatures derived from the malignant SS clusters applied to a mixed MF/SS bulk RNA cohort.24 (F) The proportion of dominant V read vs ssGSEA enrichment scores for gene signatures derived from the malignant SS clusters applied to a mixed MF/SS bulk RNA cohort.24 (G) Z-score scaled ssGSEA enrichment scores for Biocarta, Kegg, and Hallmark libraries by single-cell cluster. The median values across nonmalignant (NM) clusters are reported in a single column.
We also looked at intra and interpatient diversity using the gene expression count matrices and found the richness of malignant gene expression in malignant T cells to generally be higher than nonmalignant T cells and varied within and across patients (supplemental Figure 4). Beyond gene expression and representativeness, we also investigated potential functional differences in terms of cell cycle and signaling pathway enrichments as well as cell subset-specific signatures. Cell-cycle regression demonstrated key differences in cell-cycle assignment (P < 2.2e−16, χ2 test). C12 was highly proliferative, with 89.3% of cells in S or G2M phases (Figure 3D). In contrast, C10 was more quiescent in terms of cell cycle, with 20.6% of cells in S or G2M phases. The remaining malignant clusters have more intermediate distributions within our cell-cycle analysis (Figure 3D).
To ensure our malignant clustering was representative of CTCL T cells, we isolated highly expressed genes for each cluster to perform gene set enrichment on previously published bulk CTCL RNA-seq results (Figure 3E).24 With the normal controls, we demonstrated that C3, C6, and C12 signatures had the highest level of separation (Figure 3E). Conversely, the C10 signature had the opposite trend with lower enrichment in the SS/MF compared with normal control skin. Using the percent of dominant V-gene recovered, a surrogate marker for clonality, in the cohort, we found enrichment for C1, C2, C3, and C6 signature enrichment directly and significantly correlated with clonality (Figure 3F). Both C12 and C15 had insignificant correlations, with C12 trending toward direct correlation (Figure 3F); this may be a result of the 2 clusters representing more rare populations in CTCL. Similarly, nonmalignant T-cell clusters, C4, C7, and C8, were insignificantly correlated with the dominant V gene (supplemental Figure 5). Only C10 gene set enrichment negatively correlated with clonality (Figure 3F).
The C10 cluster was aberrant not only in global position on UMAP compared with other CTCL clusters but also lacked consistent markers of CTCL (Figure 3C). Closer examination found that of the hyperexpanded cells in C10, 63.6% of the cells in this cluster were derived from CTCL4. Next, we performed gene set enrichment across several diverse cellular processes to compare potentially functional differences between CTCL clusters and nonmalignant peripheral blood T cells (supplemental Table 4). Gene set enrichment results with column clustering based on the Manhattan distance found 3 distinct enrichment patterns. The C2-C6-C10 patterns had enrichment in Fos signaling and complement activation (Figure 3G). C10 also displayed unique enrichment in amino acid and nitrogen metabolism (Figure 3G). Nonmalignant and C1 clusters had unremarkable enrichment across pathways. The C3-C12-C15 pattern consisted of elevated enrichment of downstream signaling from cytokines (Figure 3G). For example, C15 had unique enrichment of ALK, eicosanoid, and cytokine signaling, while C12 had unique enrichment for DNA replication, epithelial-to-mesenchymal transition, and resistance to histone–deacetylase (HDAC) inhibition (Figure 3G). The latter is particularly interesting as HDAC inhibition (HDACi) represents a major treatment paradigm for CTCL.
High-resolution mRNA expression changes from HDAC inhibitor therapy
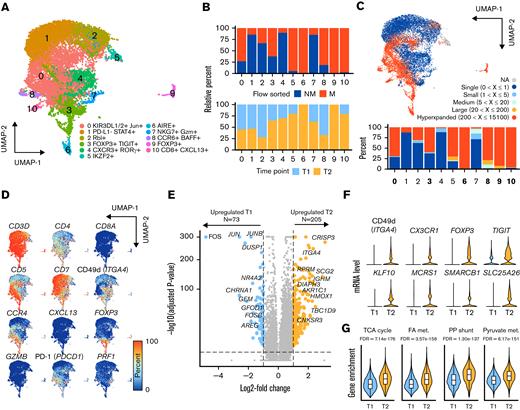
At a single-cell resolution, we have previously characterized the T-cell expression heterogeneity within a 61-year old male patient with stage IV (T4N1M0B2) SS under treatment with photopheresis and vorinostat, an HDACi. The previously published work looked at flow-sorted peripheral blood malignant (CD3+CD4+CD5brightSSChi) and nonmalignant (CD3+CD4+CD5intSSCint) T cells at an initial time point during combined HDACi and photopheresis (n = 7954) therapy. Adding to the previous data, we sequenced the same populations of peripheral blood 9 months later following disease progression (n = 7127) and integrated the single-cell sequencing runs into a single UMAP projection (Figure 4A). The result was 11 distinct clusters (Figure 4A) with malignant/nonmalignant flow-based phenotypic classifications (Figure 4B, upper panel) and distinct time points (Figure 4B, lower panel). Most clusters represented mixes of baseline (T1) and progressed (T2) time points (Figure 4B, lower panel); however, Clusters 6 and 9 were solely comprised of T cells from the T2 time point. TCR sequencing also enabled the assignment of clonotype grouping similar to the previous cohort analysis (Figure 4C). Malignant clusters 0, 3, 6, 8, 9, and 10 were defined as possessing >50% of T cells with hyperexpanded clonotypes (Figure 4C).
High-resolution mRNA expression change from HDACi therapy. (A) UMAP projection of CD3+ T-cell clusters. (B) Relative percentage of flow-sorted malignant (CD3+CD4+CD5brightSSChi) and nonmalignant (CD3+CD4+CD5intSSCint) T cells by cluster and relative percent of clusters by initial sample collection (T1) and 9 months later on HDACi therapy (T2). (C) UMAP projection with the frequency of TCR V(D)J-based clonotype attached into the following grouping: hyperexpanded (200 < X ≤ 15 100), large (20 < X ≤ 200), medium (5 < X ≤ 20), small (1 < X ≤ 5), and single copies (X = 1) using the scRepertoire R package22 with the percentage of cells in each clonotype category in the lower bar chart. (D) Percent of cells expressing selected common markers for CTCL across the UMAP projection. (E) Volcano plot of differential genes based on the comparison of T2 vs T1 malignant T cells. Significant genes, as defined by 1 > log-fold change < −1 and Bonferroni P value < .05, highlighted in colors. (F) Violin plots relative mRNA expression of selected immune, epigenetic, and resistance factor genes for T1 and T2 malignant cells. (G) Violin plots of ssGSEA enrichment of selected metabolic gene sets for T1 and T2 malignant cells.
High-resolution mRNA expression change from HDACi therapy. (A) UMAP projection of CD3+ T-cell clusters. (B) Relative percentage of flow-sorted malignant (CD3+CD4+CD5brightSSChi) and nonmalignant (CD3+CD4+CD5intSSCint) T cells by cluster and relative percent of clusters by initial sample collection (T1) and 9 months later on HDACi therapy (T2). (C) UMAP projection with the frequency of TCR V(D)J-based clonotype attached into the following grouping: hyperexpanded (200 < X ≤ 15 100), large (20 < X ≤ 200), medium (5 < X ≤ 20), small (1 < X ≤ 5), and single copies (X = 1) using the scRepertoire R package22 with the percentage of cells in each clonotype category in the lower bar chart. (D) Percent of cells expressing selected common markers for CTCL across the UMAP projection. (E) Volcano plot of differential genes based on the comparison of T2 vs T1 malignant T cells. Significant genes, as defined by 1 > log-fold change < −1 and Bonferroni P value < .05, highlighted in colors. (F) Violin plots relative mRNA expression of selected immune, epigenetic, and resistance factor genes for T1 and T2 malignant cells. (G) Violin plots of ssGSEA enrichment of selected metabolic gene sets for T1 and T2 malignant cells.
We also examined the expression distribution of common CTCL markers (Figure 4D), finding a subset of both nonmalignant and malignant T cells with expression of CD5, a feature of our patient previously described both at the mRNA level and protein level.3 Comparing the T cells with hyperexpanded clonotypes from T2 to T1, we found a total of 278 genes with significantly altered expression of adjusted P value < .05 and −1 > LFC < 1 (Figure 4E). The secondary time point had pronounced decreases across JUN/FOS genes, previously implicated in response to HDACi9 (Figure 4E). Genes that were upregulated after progression on the combined therapy were diverse and included immune mediators and transcription factors (ie, ITGA4, CXCR1, FOXP3, and TIGIT), chromatin modifiers (KLF10, MCRS1, and SMARCB1), and mitochondrial transporters (SLC25A26) (Figure 4F). A complete list of differentially regulated genes is available in supplemental Table 4. When ssGSEA was performed comparing pathway enrichment for T1 and T2, we found an overall increase in metabolic gene sets, such as fatty acid oxidation, glycolysis, and oxidative phosphorylation (Figure 4G). Conversely, we found decreased levels in gene set enrichment associated with immune ligands, such as CCR5, IL-6, and interferons (supplemental Table 5).
Defining altered expression dynamics in SS pre- and post-HDACi therapy using single cells
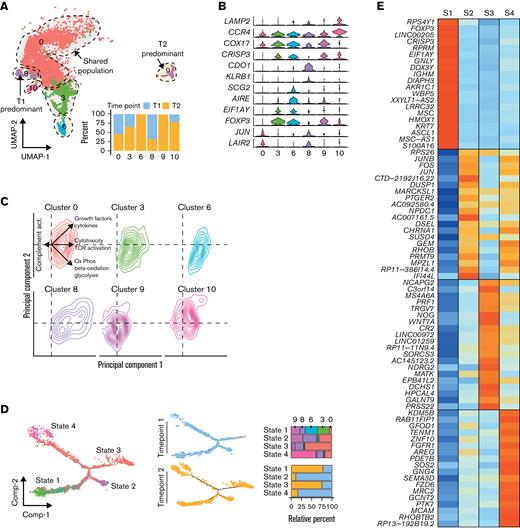
Focusing on the malignant SS cells, we defined the clusters by time point predominance, with clusters 6 and 9 new to T2 and cluster 0 (67.3% T1) and cluster 8 (72.1% T1) enriched at T1 (Figure 5A). We next attempted to characterize the gene expression differences between the clusters, finding a high degree of gene expression overlap between clusters (Figure 5B). Within Cluster 0, increased JUN and LAIR2, recently associated with HDACi resistanceField 10, were observed. With our previous characterization, we noted a single isolated cluster of FOXP3-high malignant T cells3 corresponding to Cluster 3 in our new analysis. T2 had the emergence of 2 additional clusters, 6 and 9, with moderate levels of FOXP3 expression, and cluster 10, with low levels of expression (Figure 5B). Cluster 8, the only T1-predominant cluster, had high cysteine dioxygenase type I (CDO1) gene levels. In addition to gene expression analysis, we also performed ssGSEA and examined the distribution of the malignant SS T cells and principal component analysis. Notably, the T2-predominant clusters 6 and 9 had greater representation in the lower right quadrant associated with the metabolic pathway enrichment previously mentioned (Figure 5C). In contrast, Cluster 8 had preferential enrichment in growth factor and cytokine signaling (Figure 5C).
Single-cell analysis of before and after HDACi therapy shows diverging cell populations. (A) UMAP projection of malignant T cells at initial sample collection (T1, n = 2603) and after HDACi progression (T2, n = 2778) with percent of cells in clusters by time point. (B) Violin plots of cluster-specific gene markers compared with the entire cohort of malignant T cells across both time points. (C) Density plots of the principal component analysis of ssGSEA results by malignant T-cell clusters. (D) Trajectory manifold of malignant T cells using the monocle R package with both the Seurat clusters and time point displayed. (E) Median mRNA expression of the top 20 significantly (adjusted P value < .05) upregulated markers by transition state. (F) Venn diagram of the overlap in significant genes between the transcriptional states in the cell trajectory and comparing HDACi-resistant vs -sensitive patients with SS in GSE13205310 with a selection of the 33 overlapping genes displayed as violin plots. Significant genes were defined as log-fold change ≥ 0.5 and adjusted P value < .05. (G) Gene expression patterns of significantly expressed genes by transition state that are previously reported upregulated in an HDACi-resistant SS10 along the cell trajectory manifold.
Single-cell analysis of before and after HDACi therapy shows diverging cell populations. (A) UMAP projection of malignant T cells at initial sample collection (T1, n = 2603) and after HDACi progression (T2, n = 2778) with percent of cells in clusters by time point. (B) Violin plots of cluster-specific gene markers compared with the entire cohort of malignant T cells across both time points. (C) Density plots of the principal component analysis of ssGSEA results by malignant T-cell clusters. (D) Trajectory manifold of malignant T cells using the monocle R package with both the Seurat clusters and time point displayed. (E) Median mRNA expression of the top 20 significantly (adjusted P value < .05) upregulated markers by transition state. (F) Venn diagram of the overlap in significant genes between the transcriptional states in the cell trajectory and comparing HDACi-resistant vs -sensitive patients with SS in GSE13205310 with a selection of the 33 overlapping genes displayed as violin plots. Significant genes were defined as log-fold change ≥ 0.5 and adjusted P value < .05. (G) Gene expression patterns of significantly expressed genes by transition state that are previously reported upregulated in an HDACi-resistant SS10 along the cell trajectory manifold.
In order to better understand the transition of the malignant SS T cells at baseline and following progression on HDACi therapy, we used reverse-graph embedding15 to construct a cellular trajectory manifold that can identify distinct cellular fates or transcriptional states (Figure 5D). Defining each branch of the manifold as transcriptional states, we found 4 distinct states with a clear separation in relative percent of cells by time point. The T1 states were defined as State 2 and State 4, while State 1 and State 3 were formed from the majority of T2 malignant SS cells (Figure 5D). Differences were also noted in the placement of time point cells along the manifold with T2 cells of the State 3 and State 4 branches at the distal portions compared with the more proximal T1 cells. We performed differential gene expression analysis for each transcriptional state, comparing the state to the rest of the cells (Figure 5E). Most interestingly, State 1, comprised of the FOXP3+ clusters 3, 6, and 9, was a T2-predominant transcriptional state with multiple clusters with FOXP3, potentially implicating a therapy-induced genetic program. We next wanted to understand how the branches of the manifold contribute to the resistance to therapy. Using recent gene expression results for patients sensitive and resistant to HDACi,10 we compared differential gene expression between all 4 states and found a total of 33 genes previously reported (Figure 5F). Of the 1395 genes that were altered that were not previously reported, the preponderance of the most significant genes was increased in the State 1 cells (Figure 5G). It should be noted that these genes may represent new mechanisms of resistance, underlying genetic changes induced by HDACi, or a combination of the two. Toward understanding the functional import of these genes, we performed gene set enrichment analysis (supplemental Figure 6). Among the most significantly enriched gene sets were amino and fatty acid metabolism, DNA repair pathways, and cytokine signaling.
Discussion
Despite being a clonal T-cell neoplasm, we and others have reported that a degree of epigenomic, transcriptional, and protein heterogeneity exists in both SS and MF.3-5,34,35 Unlike previous single-cell quantifications of CTCL, we integrated our transcriptomic data across patients, allowing for the quantification of CTCL heterogeneity across patients. Indeed, leveraging the single-cell mRNA and TCR sequencing of circulating nonmalignant and SS cells for assessment of clonality, we find a high degree of inter and intrapatient heterogeneity (Figures 2 and 3) across over 50 000 cells. This variability may contribute to the discordance in the clinical or histopathological definition of CTCL, prolonged median time to diagnosis of 3 years, and therapeutic treadmilling observed in patient management.36,37 A new report has even shown distinct transcriptional signatures of skin-derived and blood-derived MF cells at the level of single patients.38 Despite this heterogeneity, we found common SS markers that may warrant further investigation, including several previously identified, such as NEDD4L,29,39PLS3,40 and TOX.41,42 Notably, the AIRE gene was expressed across samples in 58% of malignant cells vs 8.7% of nonmalignant cells. The largest difference for non-TCR–related genes and only recently AIRE expression was incidentally reported in circulating mycosis fungoides cells.43 The autoimmune regulator, AIRE, functions in central tolerance during T-cell maturation and is expressed by thymic epithelial and dendritic cells.44 Loss of AIRE leads to autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, a pleiotropic genetic immune dysfunction characterized by the loss of thymus-generated Tregs.45 The role of AIRE in the ectopic expression of peripheral antigens raises the possibility that its expression by CTCL cells may function to recruit and stimulate a wider array of Tregs, potentially as a mechanism to avoid immune responses.
Within the CTCL literature, there is a lack of consensus regarding the existence of a FOXP3+ regulatory or regulatory-like phenotype of malignant T cells.46-49 This variation in the literature may be a result of the regulation of FOXP3 expression by environmental factors, such as bacterial toxin50 or therapeutic regimens. In terms of the latter, the use of HDACi led to the development of FOXP3-moderate–expressing clusters (labeled 6 and 9 in Figure 5F). This is underscored by previous findings from a single-cell Assay for Transposase-Accessible Chromatin (ATAC)-seq study, which reported differential accessibility of FOXP3 in clonal SS cells.35 Upregulation of HDAC9 has been previously reported in CTCL,9,51 which along with HDAC6, has been shown to regulate the expression of FOXP3 in Tregs.52 A subset of HDACi-responsive patients with CTCL had an opening of the chromatin associated with the FOXP3 enhancers, suggesting an increase in FOXP3 expression may be a surrogate marker of response to HDAC inhibition.9
We previously showed that FOXP3 was the single strongest predictor of early-stage disease in a cohort of 152 patients with CTCL.3,53,54 With transcriptional State 1, a post-HDACi–predominant state consisting of multiple populations of FOXP3+ malignant SS cells, further research is warranted to distinguish whether therapy selects for diverging populations of FOXP3+ or pushes the transcriptional state to that of earlier-stage disease. The latter has been suggested as HDACi increases chromatin access to the FOXP3 loci.9 However, 2 major caveats to this concept include (1) the shift in the transcriptional state may depend on the specific HDACi agent, and (2) further work is needed to understand if this is truly a transcriptional state vs epigenomic shift. To the former, vorinostat inhibits HDAC class I, II (to include HDAC9), and IV, while the potent bicyclic romidepsin is more specific for HDAC class I molecules. Key differences in HDACi agent-specific effects on genome accessibility, with romidepsin preferentially altering promoters and active enhancer regions compared with vorinostat, likely alters gene expression.9 This differential effect of HDACi on heterogeneity via flow cytometry has been previously reported with ex vivo treatment of SS with vorinostat and romidepsin.4
Acknowledgments
The authors thank Julie McKillip for her excellent nursing support.
Research reported in this publication was supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under Award Number P30CA086862, Public Health Service grant number P50 CA097274 from the University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence (UI/MC Lymphoma SPORE) and the NCI, and a Cutaneous Lymphoma Catalyst Research Grant from the Cutaneous Lymphoma Foundation. The data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility at the University of Iowa, funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran's Administration Medical Center, as well as at the Genomics Division of the Iowa Institute of Human Genetics, which is supported, in part, by the University of Iowa Carver College of Medicine and the Holden Comprehensive Cancer Center (NCI of the NIH under Award Number P30CA086862). Salary support was provided from the NIH under a National Institute of Arthritis and Musculoskeletal and Skin K08 award AR069111 (Principal Investigator: A.J.), VA Merit Award I01 BX004907, and an NCI NIH F30 fellowship CA206255 (Principal Investigator: N.B.).
Authorship
Contribution: N.B. processed and analyzed the data and wrote the manuscript; K.J.S., N.H., and L.S.O. collected and processed clinical samples; A.C.R. provided clinical insight and assisted in recruiting patients; A.M.B. and V.L. performed immunohistochemical staining and quantification, provided clinical insight, and assisted in drafting the manuscript; B.K.L. provided clinical insight and assisted in drafting the manuscript; and A.R.M. and A.J. designed research, assisted in drafting the manuscript, and supervised the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ali Jabbari, University of Iowa, 2110A Medical Laboratories, 500 Newton Rd, Iowa City, IA 52242; e-mail: ali-jabbari@uiowa.edu.
References
Author notes
Raw and processed data are available at GEO (accession numbers GSE124899 and GSE146586). Please direct other inquiries to the corresponding author, Ali Jabbari (ali-jabbari@uiowa.edu).
The full-text version of this article contains a data supplement.


![Single-cell map of peripheral blood across 6 patients with Sézary syndrome. (A) UMAP projection of the flow-sorted CD45+ immune cells (n = 54 994) across 6 patients: T cells/CTCL cells (Cluster 1, 2, 3, 4, 6, 7, 8, 9, 10, 12, and 15), myeloid cells (Cluster 5, 11, and 14), and B cells (Cluster 13). (B) Percent of cells expressing selected lineage markers for T cells (CD2 and CD3G), myeloid cells (ITGAM [CD11b] and ITGAX [CD11c]), and B cells (CD19 and CD79A). (C) Normalized correlation values across rows ranging from 0 to 1 for predicted immune cell phenotypes from the Human Primary Cell Atlas18 based on the SingleR R package17 for each cluster. (D) Percentage of total cells for each patient by cell type. (E) Cell distribution by patient sample both as contour graphs and summarized in the accompanying bar chart by patient and cluster. Clusters representing >15% of total patient cells are labeled.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/3/10.1182_bloodadvances.2021005991/3/m_blooda_adv-2021-005991-gr1.jpeg?Expires=1767721290&Signature=du3C-5UVjX1rlJmxgmaK7qInWudYFuhLOcdl3RhKXWnT6S~m3vjDcINBcUtREajB0Z3iyyGTUxDgtSsPkfuoq0QGbgFDV~L9f2VSROTcwhyTNRpeYzWp2H38xoKn9~RtVjSnjt4HsWM2vf-h-YbDkzCbjfdJEFuy6dlSOKMIjFw86WpVB3GeTaOjymxBXUPYq4yTELCcx5Fcwzg7Hg2IN6DG81buHbQzCnP9ukfiac-0Nl5cWuWHq2kM7~tyI8qmzjEGf5pb-QBzVJQixge2wPg6P41rIZiCtJqQdU3Zl7822dwA755GUCFu5KJzEBnez3HuKcfjnrEc~0FkOeDwgA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)