Using proteomics, diagnostic FL samples showed different protein profiles according to risk of subsequent high-grade transformation.
Protein expression of CASP3, MCL1, BAX, BCL-xL, and BCL-rambo suggest apoptotic deregulation in FL predictive of subsequent transformation.
Visual Abstract
Follicular lymphoma (FL) is an indolent lymphoma with a generally favorable prognosis. However, histological transformation (HT) to a more aggressive disease leads to markedly inferior outcomes. This study aims to identify biological differences predictive of HT at the time of initial FL diagnosis. We show differential protein expression between diagnostic lymphoma samples from patients with subsequent HT (subsequently-transforming FL [st-FL]; n = 20) and patients without HT (nontransforming FL [nt-FL]; n = 34) by label-free quantification nano liquid chromatography-tandem mass spectrometry analysis. Protein profiles identified patients with high risk of HT. This was accompanied by disturbances in cellular pathways influencing apoptosis, the cytoskeleton, cell cycle, and immune processes. Comparisons between diagnostic st-FL samples and paired transformed FL (n = 20) samples demonstrated differential protein profiles and disrupted cellular pathways, indicating striking biological differences from the time of diagnosis up to HT. Immunohistochemical analysis of apoptotic proteins, CASP3, MCL1, BAX, BCL-xL, and BCL-rambo, confirmed higher expression levels in st-FL than in nt-FL samples (P < .001, P = .015, P = .003, P = .025, and P = .057, respectively). Moreover, all 5 markers were associated with shorter transformation-free survival (TFS; P < .001, P = .002, P < .001, P = .069, and P = .010, respectively). Notably, combining the expression of these proteins in a risk score revealed increasingly inferior TFS with an increasing number of positive markers. In conclusion, proteomics identified altered protein expression profiles (particularly apoptotic proteins) at the time of FL diagnosis, which predicted later transformation.
Introduction
Follicular lymphoma (FL), the second most common lymphoma entity in Western countries, is an indolent disease that is generally considered incurable. The lymphoma arises from germinal center B cells, in which particularly BCL2 gene rearrangements represent a diagnostic hallmark.1,2 Over the past 2 decades, the outcome for patients with FL has improved considerably.3,4 However, despite the generally favorable prognosis, patients at high risk, especially those undergoing histological transformation (HT) to a more aggressive lymphoma entity, represent a group with limited treatment options, often associated with a dismal outcome. Transformation is associated with the development of a more aggressive looking histology, most often corresponding to diffuse large B-cell lymphoma (DLBCL). To date, no single biological event has been proven specific for HT; however, studies have highlighted recurrent aberrations in genes with roles in the dysregulation of cell cycle control and DNA damage response, providing biological clues to an understanding of the pathogenesis of HT.5-11
Proteomics is a valuable tool for large-scale analysis of complex cell systems such as neoplasms. By deciphering cellular mechanisms at the protein level, important insights can be gained into cancer pathogenesis that may not be accessible using other (eg, RNA-based) technologies.12 Moreover, analysis of the proteome has the potential to capture novel aspects of tumor heterogeneity, contributing to biomarker discovery and providing candidates for novel targeted therapies in modern precision medicine.13,14 Given that the proteome is adaptive and changes in response to various factors acting at different biological levels,15-17 a better understanding of the proteome in FL could provide key insights into the biological mechanisms that drive transformation in FL.14,18
To investigate FL protein profiles in the context of high-risk disease with HT as the end point, we studied diagnostic lymphoma samples using mass spectrometry (MS). Using this proteomics-based assessment, we identified differentially expressed proteins in diagnostic FL samples from patients with and those without subsequent transformation.
Patients and methods
Patient samples
Analyses were performed on diagnostic formalin-fixed, paraffin-embedded lymphoma specimens from 54 patients diagnosed with FL grade 1 to 3A at Aarhus University Hospital, Denmark between 1990 and 2015.19-21 These included 34 patients with FL without HT with at least 10 years of follow-up (nontransforming FL [nt-FL]) and 20 patients with FL with a subsequent histologically confirmed transformation (subsequently-transforming FL [st-FL]) to DLBCL or FL grade 3B, at least 6 months after the primary diagnosis. Paired high-grade lymphoma samples from the time of HT were also analyzed (histologically transformed FL [tFL]; n = 20). All biopsies were reviewed by 2 experienced hematopathologists (S.J.H.-D. and T.L.P.) and classified according to the 2017 update to the World Health Organization classification.2 Clinicopathological data were collected from the Danish Lymphoma Registry22,23 and have been described previously.19-21,24,25
The study was approved by the Danish National Committee on Health Research Ethics (1-10-72-276-13) and the Danish Data Protection Agency (1-16-02-407-13) and was conducted in accordance with the Declaration of Helsinki.
Identification of differentially expressed proteins
To identify differentially expressed proteins between the samples, a label-free quantification nano liquid chromatography-tandem MS–based proteomic analysis was performed.26,27 The procedure is described in detail in the supplemental Methods. In brief, proteins from formalin-fixed, paraffin-embedded lymphoma tissues were extracted and proteolytically digested into peptides.16,27 Peptides were separated by nano liquid chromatography and analyzed in the mass spectrometer.16 Bioinformatic analysis was performed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database and with the use of Qiagen Ingenuity Pathway Analysis (IPA) (Qiagen Inc).28
Immunohistochemical staining of selected proteins
Selected differentially expressed proteins identified by MS-based proteomics were evaluated using immunohistochemistry. These included caspase 3 (CASP3), induced myeloid leukemia cell differentiation protein (MCL1), BCL-2–associated X protein (BAX), BCL-extralarge (BCL-xL), and BCL2-like 13 (BCL-rambo). A detailed description of staining protocols is given in the supplemental Methods and supplemental Table 1. Staining was quantified by digital image analysis. The quantification outputs were area fractions, defined as the stained area normalized to the total area within the region of interest.20,21,29 Expression levels of CASP3, MCL1, BAX, and BCL-xL were based on all positive staining, whereas expression levels of BCL-rambo were based on strong-intensity staining.
Statistical analysis
Differences in clinicopathological features were assessed using a χ2 test or Fisher exact test. Student t test was used for statistical analysis of fold changes of differentially expressed proteins in the MS-based proteomics analysis. Principal component analyses (PCAs) were performed on proteins with no missing values to avoid imputation. Hierarchal clustering was performed with Euclidean distance as the dissimilarity measure, and Ward linkage was used to join clusters. Differences in staining area fractions among nt-FL, st-FL, and tFL samples were assessed using an independent Mann-Whitney U test and a paired Wilcoxon ranked sum test. Correlation of biomarker expression and clinicopathological features was evaluated using a Spearman rank test. Time-related end points were analyzed using the Kaplan-Meier and log rank method, with transformation-free survival (TFS) as the end point. TFS was defined as the time from initial FL diagnosis to the date of transformation.19,21 Cutoff values for high vs low expression of all 5 biomarkers and TFS analyses were determined by a receiver operating characteristic (curve) analysis, with the optimal cutoff point calculated using Youden index. P values < .05 were considered statistically significant. Statistical analyses were performed using R statistical software (version 4.1.2).
Results
Patients’ characteristics
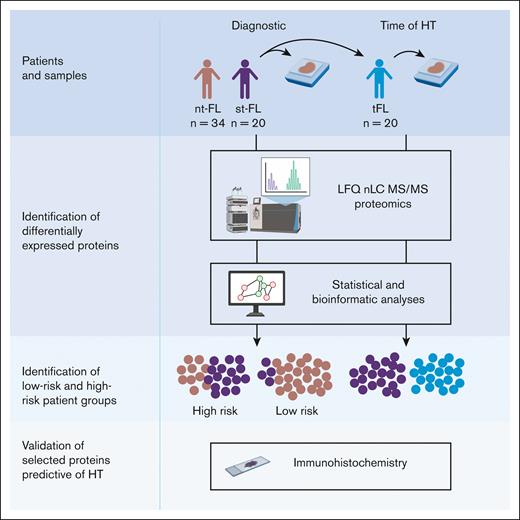
The patient cohort comprised 54 patients with FL, including 20 patients with st-FL with subsequent HT and 34 patients with nt-FL without (Table 1). The study included an equal number of males and females, and ages ranged from 35 to 78 years, with a median age of 54 years at FL diagnosis. Patients with subsequent HT had slightly worse risk profiles than those with nt-FL, with more advanced Ann Arbor stage and, thus, higher follicular lymphoma international prognostic index (FLIPI) risk scores. No significant differences were found in other common clinicopathological features. For the patients with st-FL, in addition to the diagnostic FL sample, a paired high-grade biopsy from the time of HT was included to identify HT-related differences in protein expression (Figure 1).
Patients’ clinicopathological features
| Characteristics . | All, N = 54 n (%) . | nt-FL, n = 34 n (%) . | st-FL, n = 20 n (%) . | P . |
|---|---|---|---|---|
| Sex | NS | |||
| Male | 27 (50) | 16 (47) | 11 (55) | |
| Female | 27 (50) | 18 (53) | 9 (45) | |
| Age at FL diagnosis, y | NS | |||
| Median | 54 | 54 | 54 | |
| Range | 35-78 | 35-76 | 40-78 | |
| Ann Arbor stage | .014 | |||
| I-II | 17 (26) | 15 (44) | 2 (10) | |
| III-IV | 35 (71) | 18 (53) | 17 (85) | |
| Unknown | 2 (3) | 1 (3) | 1 (5) | |
| FLIPI | .012 | |||
| Low | 24 (37) | 20 (59) | 4 (20) | |
| Intermediate | 19 (28) | 10 (29) | 9 (45) | |
| High | 7 (29) | 2 (6) | 5 (25) | |
| Unknown | 4 (6) | 2 (6) | 2 (10) | |
| LDH elevation | NS | |||
| Yes | 2 (12) | 1 (3) | 1 (5) | |
| No | 48 (82) | 31 (91) | 17 (85) | |
| Unknown | 4 (6) | 2 (6) | 2 (10) | |
| B-symptoms | NS | |||
| Yes | 12 (23) | 6 (18) | 6 (30) | |
| No | 39 (72) | 27 (79) | 12 (60) | |
| Unknown | 3 (6) | 1 (3) | 2 (10) | |
| Performance score | NS | |||
| <2 | 41 (75) | 28 (82) | 13 (65) | |
| ≥2 | 10 (20) | 5 (15) | 5 (25) | |
| Unknown | 3 (5) | 1 (3) | 2 (10) | |
| Bone marrow involvement | NS | |||
| Yes | 14 (31) | 6 (18) | 8 (40) | |
| No | 33 (55) | 24 (71) | 9 (45) | |
| Unknown | 7 (14) | 4 (12) | 3 (15) | |
| Anemia | NS | |||
| Yes | 4 (8) | 1 (3) | 3 (15) | |
| No | 47 (88) | 32 (94) | 15 (75) | |
| Unknown | 3 (5) | 1 (3) | 2 (10) | |
| FL histology | NS | |||
| FL grade 1-2 | 46 (86) | 29 (85) | 17 (85) | |
| FL grade 3A | 8 (14) | 5 (15) | 3 (15) |
| Characteristics . | All, N = 54 n (%) . | nt-FL, n = 34 n (%) . | st-FL, n = 20 n (%) . | P . |
|---|---|---|---|---|
| Sex | NS | |||
| Male | 27 (50) | 16 (47) | 11 (55) | |
| Female | 27 (50) | 18 (53) | 9 (45) | |
| Age at FL diagnosis, y | NS | |||
| Median | 54 | 54 | 54 | |
| Range | 35-78 | 35-76 | 40-78 | |
| Ann Arbor stage | .014 | |||
| I-II | 17 (26) | 15 (44) | 2 (10) | |
| III-IV | 35 (71) | 18 (53) | 17 (85) | |
| Unknown | 2 (3) | 1 (3) | 1 (5) | |
| FLIPI | .012 | |||
| Low | 24 (37) | 20 (59) | 4 (20) | |
| Intermediate | 19 (28) | 10 (29) | 9 (45) | |
| High | 7 (29) | 2 (6) | 5 (25) | |
| Unknown | 4 (6) | 2 (6) | 2 (10) | |
| LDH elevation | NS | |||
| Yes | 2 (12) | 1 (3) | 1 (5) | |
| No | 48 (82) | 31 (91) | 17 (85) | |
| Unknown | 4 (6) | 2 (6) | 2 (10) | |
| B-symptoms | NS | |||
| Yes | 12 (23) | 6 (18) | 6 (30) | |
| No | 39 (72) | 27 (79) | 12 (60) | |
| Unknown | 3 (6) | 1 (3) | 2 (10) | |
| Performance score | NS | |||
| <2 | 41 (75) | 28 (82) | 13 (65) | |
| ≥2 | 10 (20) | 5 (15) | 5 (25) | |
| Unknown | 3 (5) | 1 (3) | 2 (10) | |
| Bone marrow involvement | NS | |||
| Yes | 14 (31) | 6 (18) | 8 (40) | |
| No | 33 (55) | 24 (71) | 9 (45) | |
| Unknown | 7 (14) | 4 (12) | 3 (15) | |
| Anemia | NS | |||
| Yes | 4 (8) | 1 (3) | 3 (15) | |
| No | 47 (88) | 32 (94) | 15 (75) | |
| Unknown | 3 (5) | 1 (3) | 2 (10) | |
| FL histology | NS | |||
| FL grade 1-2 | 46 (86) | 29 (85) | 17 (85) | |
| FL grade 3A | 8 (14) | 5 (15) | 3 (15) |
P values in bold are significant. FLIPI, follicular lymphoma international prognostic index; LDH, lactate dehydrogenase; NS, not significant.
Study workflow diagnostic samples from nt-FL and st-FL samples were analyzed by large-scale MS-based proteomics to investigate protein expression differences between the 2 patient groups. In addition, for the transforming FL group, a paired high-grade sample from the time of HT was also analyzed. Subsequent statistical and bioinformatic analyses were performed to identify risk groups based on the respective protein expression profiles. Lastly, selected proteins were evaluated using targeted immunohistochemistry. LFQ, label-free quantification. Created with BioRender.com.
Study workflow diagnostic samples from nt-FL and st-FL samples were analyzed by large-scale MS-based proteomics to investigate protein expression differences between the 2 patient groups. In addition, for the transforming FL group, a paired high-grade sample from the time of HT was also analyzed. Subsequent statistical and bioinformatic analyses were performed to identify risk groups based on the respective protein expression profiles. Lastly, selected proteins were evaluated using targeted immunohistochemistry. LFQ, label-free quantification. Created with BioRender.com.
Proteomic profiling identifies patients with high-risk FL
The protein composition of all included samples was assessed by liquid chromatography-tandem MS–based proteomics. A combined set of 2665 proteins were identified across the 3 patient groups (supplemental Table 2).
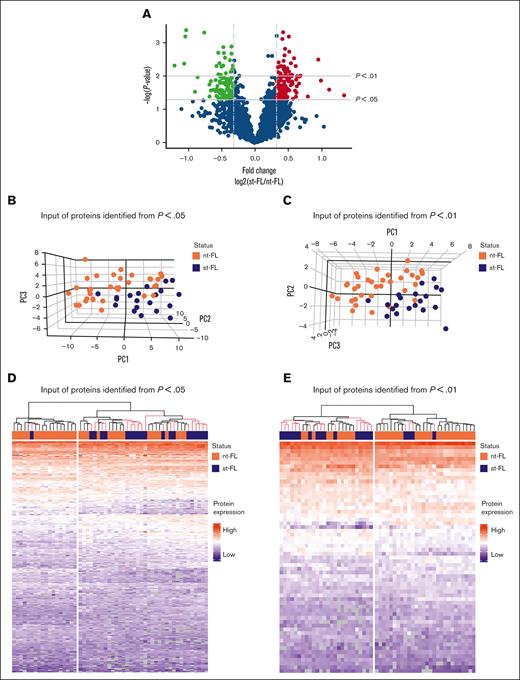
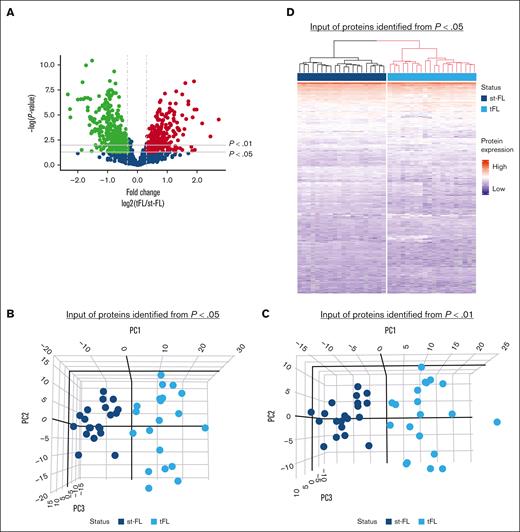
Comparing diagnostic nt-FL and st-FL samples, 795 proteins were identified with at least 20% difference in fold changes (fold changes, 0.43-2.51; supplemental Table 3; Figure 2A). Among these, 242 proteins were significantly differentially expressed (P < .05), including 138 proteins that were upregulated in st-FL samples (fold changes, 1.20-2.51) and 104 proteins that were downregulated (fold changes, 0.43-0.84; supplemental Table 4). Among significantly upregulated proteins were matrix metalloproteinase 9, CASP3, BAX, MCL1, BCL-xL, BCL-rambo, cell division cycle 26, myeloperoxidase, and PAX5, whereas significantly downregulated proteins included MEK1, proto-oncogene GTPase, β-actin, CD81, and phosphoinositide-3-kinase adapter protein 1. To better describe the most significant differences between patient groups, a threshold of P < .01 was also applied, resulting in 61 differentially expressed proteins at this setting.
Differentially expressed proteins identified by MS-based proteomics differentiates, already at diagnosis, FL with and without subsequent transformation. (A) Proteins identified in diagnostic nt-FL and st-FL samples. The x-axis represents the fold changes as transformed by log2(st-FL/nt-FL); thus, red dots mark upregulated proteins at P < .05 and a fold change >20%, whereas green dots mark downregulated proteins at P < .05 and a fold change <20%. To allow for better visualization of highly significant data points, the negative logarithm (log10) of the P values are plotted on the y-axis. Horizontal lines mark P values of P < .05 and P < .01, respectively. Vertical dashed lines mark proteins with fold changes of at least 20%. (B) 3D PCA plot with input of differentially expressed proteins at P < .05 comparing nt-FL and st-FL samples. (C) 3D PCA plot with input of differentially expressed proteins at P < .01 comparing nt-FL and st-FL samples. (D) Hierarchal clustering analysis based on differentially expressed proteins at P < .05 comparing nt-FL and st-FL samples. (E) Hierarchal clustering analysis based on differentially expressed proteins at P < .01 between nt-FL and st-FL samples.
Differentially expressed proteins identified by MS-based proteomics differentiates, already at diagnosis, FL with and without subsequent transformation. (A) Proteins identified in diagnostic nt-FL and st-FL samples. The x-axis represents the fold changes as transformed by log2(st-FL/nt-FL); thus, red dots mark upregulated proteins at P < .05 and a fold change >20%, whereas green dots mark downregulated proteins at P < .05 and a fold change <20%. To allow for better visualization of highly significant data points, the negative logarithm (log10) of the P values are plotted on the y-axis. Horizontal lines mark P values of P < .05 and P < .01, respectively. Vertical dashed lines mark proteins with fold changes of at least 20%. (B) 3D PCA plot with input of differentially expressed proteins at P < .05 comparing nt-FL and st-FL samples. (C) 3D PCA plot with input of differentially expressed proteins at P < .01 comparing nt-FL and st-FL samples. (D) Hierarchal clustering analysis based on differentially expressed proteins at P < .05 comparing nt-FL and st-FL samples. (E) Hierarchal clustering analysis based on differentially expressed proteins at P < .01 between nt-FL and st-FL samples.
To determine whether different protein expression profiles could differentiate FL samples according to subsequent risk of transformation, unsupervised PCAs were performed. No clear separation was present when based on the combined set of identified proteins (supplemental Figure 1). However, focusing on significantly differentially expressed proteins (at P < .05 or P < .01, respectively) revealed a strong pattern of samples corresponding to nt-FL and st-FL samples, reflecting possible risk subgroups of FL tumors with HT as the end point (Figure 2B-C).
Studying the protein profiles in the tumor samples, hierarchal clustering based on 242 significantly differentially expressed proteins (P < .05), identified 2 groups: (1) again a low-risk group (17 nt-FL and 1 st-FL) and (2) a high-risk group (17 nt-FL and 19 st-FL; further designated as analysis A; Figure 2D). Finally, with input from 61 differentially expressed proteins identified at the P < .01 threshold, this separation was not improved but showed a more mixed grouping of nt-FL and st-FL, that is: (1) a low-risk group (24 nt-FL and 4 st-FL) and (2) a high-risk group (10 nt-FL and 16 st-FL; further designated analysis B; Figure 2E).
High-risk groups show separation according to protein profiles
Analysis A and B both revealed high-risk clusters of nt-FL and st-FL samples; samples within the respective subgroups were then reanalyzed to identify candidate proteins with even higher potential for predicting subsequent HT.
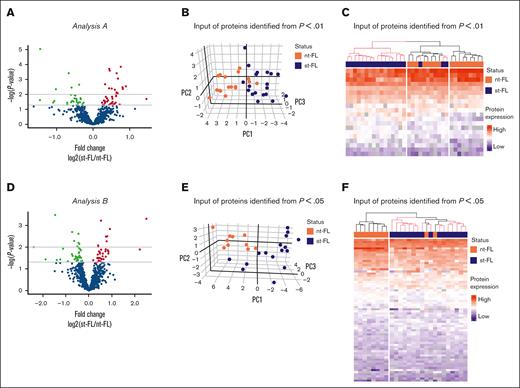
In analysis A, the high-risk group of 17 nt-FL and 19 st-FL samples was reanalyzed. Here, 63 proteins were identified as significantly differentially expressed at P < .05 between the nt-FL and st-FL high-risk samples, with 20 proteins differentially expressed at P < .01 (Figure 3A; supplemental Table 6). PCA with input of proteins differentially expressed at P < .05 was able to further discriminate nt-FL and st-FL samples in the high-risk subgroup (supplemental Figure 2A-B). This observation was even more evident with an input of proteins differentially expressed at P < .01, which revealed a separation with only a few samples remaining unclassified intermingled (Figure 3B). This was also reflected by hierarchal clustering, in which an input of 20 differentially expressed proteins at P < .01 revealed a clear high-risk cluster consisting of only st-FL samples (Figure 3C). Notably, in the corresponding low-risk cluster, 3 st-FL samples were included among nt-FL samples. Clinicopathological data showed no distinct explanation for the placement of these 3 st-FL samples; however, the patients were all younger (age, 47-57 years) and diagnosed with FL grade 2, stage IV. Two patients experienced HT ∼1 year after initial FL diagnosis, whereas the remaining patient experienced HT after 6 years.
Reanalysis of patients with high-risk FL from analysis A and analysis B. (A) Identified proteins comparing nt-FL and st-FL samples from the high-risk group identified in analysis A. The x-axis represents the fold changes as transformed by log2(st-FL/nt-FL); thus, red dots mark upregulated proteins at P < .05 and a fold change >20%, whereas green dots mark downregulated proteins at P < .05 and a fold change <20%. To allow for better visualization of highly significant data points, the negative logarithm (log10) of the P values are plotted on the y-axis. Horizontal lines mark P values of P < .05 and P < .01, respectively. (B) 3D PCA based on differentially expressed proteins at P < .01 identified in high-risk group in analysis A. (C) Hierarchal clustering based on differentially expressed proteins at P < .01 identified in the high-risk group from analysis A. (D) Identified proteins comparing nt-FL and st-FL samples from the high-risk group identified from analysis B. Fold change as transformed by log2(st-FL/nt-FL). Red, upregulated at P < .05 threshold; green, downregulated at P < .05 threshold. Horizontal lines mark P values of P < .05 and P < .01, respectively. (E) 3D PCA based on differentially expressed proteins at P < .05 in the high-risk group from analysis B. (F) Hierarchal clustering based on differentially expressed proteins at P < .05 in the high-risk group from analysis B.
Reanalysis of patients with high-risk FL from analysis A and analysis B. (A) Identified proteins comparing nt-FL and st-FL samples from the high-risk group identified in analysis A. The x-axis represents the fold changes as transformed by log2(st-FL/nt-FL); thus, red dots mark upregulated proteins at P < .05 and a fold change >20%, whereas green dots mark downregulated proteins at P < .05 and a fold change <20%. To allow for better visualization of highly significant data points, the negative logarithm (log10) of the P values are plotted on the y-axis. Horizontal lines mark P values of P < .05 and P < .01, respectively. (B) 3D PCA based on differentially expressed proteins at P < .01 identified in high-risk group in analysis A. (C) Hierarchal clustering based on differentially expressed proteins at P < .01 identified in the high-risk group from analysis A. (D) Identified proteins comparing nt-FL and st-FL samples from the high-risk group identified from analysis B. Fold change as transformed by log2(st-FL/nt-FL). Red, upregulated at P < .05 threshold; green, downregulated at P < .05 threshold. Horizontal lines mark P values of P < .05 and P < .01, respectively. (E) 3D PCA based on differentially expressed proteins at P < .05 in the high-risk group from analysis B. (F) Hierarchal clustering based on differentially expressed proteins at P < .05 in the high-risk group from analysis B.
In analysis B, reanalysis of the high-risk group of 10 nt-FL and 16 st-FL samples resulted in 68 significantly differentially expressed proteins at P < .05 and 16 differentially expressed proteins at the P < .01 threshold (Figure 3D; supplemental Table 7). Of particular note was an analysis of the 68 differentially expressed proteins at P < .05, which revealed a separation of an absolute low-risk group as well as a high-risk group containing only 2 nt-FL samples (Figure 3E-F). These 2 patients with nt-FL were a 55-year-old male and a 59-year-old female, both diagnosed with FL grade 1. Interestingly, both patients had progressive, relapsing disease, which might explain their clustering with the high-risk disease group. Similar results were seen with an input of 16 differentially expressed proteins at P < .01, which allowed for an almost complete separation of the nt-FL and st-FL samples (supplemental Figure 2C-D).
Interestingly, the same 1 st-FL sample was clustered into low-risk groups in all analyses (Figure 2D-E). This specific patient was a 40-year-old male, who presented with FL grade 1, stage III. Otherwise, clinicopathological data showed no adverse factors associated with aggressive disease at diagnosis. However, particularly noteworthy, from the time of initial FL diagnosis, 11 years passed before the patient experienced HT. This raises the possibility that the late transformation could have allowed for the accumulation of oncogenic or refractory mutations in the tumor. Furthermore, it highlights the question whether HT is an inevitable end point in all FL, including those presenting with low-grade, low-risk disease at diagnosis.
Protein profiles reveal disturbed biological pathways depending on subsequent transformation status
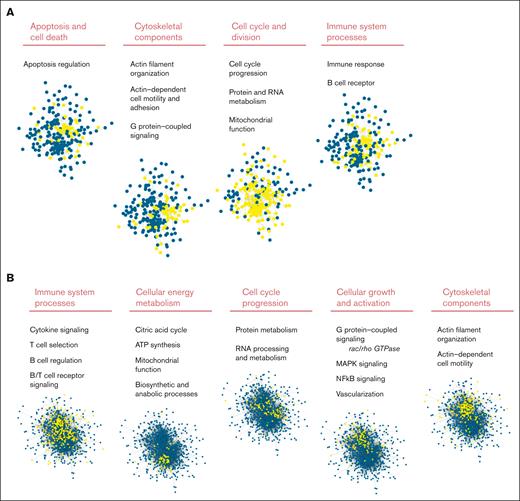
Gene enrichment analysis identified disturbed cellular pathways comparing tumor biopsy specimen from patients with nt-FL and from those with st-FL (Figure 4A). With 241 nodes and 808 edges, the protein network had significantly more interactions than expected, at random (P < .001). Different clusters of protein involvement were observed among the perturbed pathways, with many proteins involved in multiple pathways, thereby connecting the cellular pathways in several ways (Figure 4A; supplemental Table 8). Interestingly, the pathway analyses revealed striking biological differences even though most changes in protein expression were subtle (most changes were less than twofold). Especially noticeable changes were observed in the processes of cell cycle division, the cytoskeleton, apoptotic signaling, and cell death as well as the immune system (Figure 4A). Taken together, this suggests that biological differences may be present in the tumors that ultimately influence cell growth and survival. Furthermore, affected immune system processes could indicate important differences in the nonmalignant tumor microenvironment (TME). Analyses based on the IPA software with input of the combined set of differentially expressed proteins showed virtually similarly perturbed pathways with emphasis on pathways of apoptosis, molecular mechanisms of cancer, and family small GTPase (RAC) signaling (supplemental Tables 9 and 10).
STRING analysis of significantly differentially expressed proteins comparing st-FL and nt-FL samples as well as st-FL and tFL samples. Different significantly disturbed cellular pathways identified in the STRING analysis based on (A) input of 242 proteins significantly differentially expressed at P < .05 comparing nt-FL and st-FL samples. The groups contain proteins belonging to apoptotic signaling, the cytoskeleton, cell cycle signaling, and the immune system. (B) Input of the 800 proteins significantly differentially expressed at P < .05 comparing tFL and st-FL samples. The groups contain proteins belonging to the immune system, energy metabolism, cell cycle, cellular growth and activation, and the cytoskeleton. Nodes represent proteins and edges visualize interactions. The different cellular pathways visualized were significantly disturbed in the STRING analysis. Yellow indicates proteins involved in said pathway; blue, protein not involved in the pathway. ATP, adenosine triphosphate; GTPase, guanosine triphosphatase.
STRING analysis of significantly differentially expressed proteins comparing st-FL and nt-FL samples as well as st-FL and tFL samples. Different significantly disturbed cellular pathways identified in the STRING analysis based on (A) input of 242 proteins significantly differentially expressed at P < .05 comparing nt-FL and st-FL samples. The groups contain proteins belonging to apoptotic signaling, the cytoskeleton, cell cycle signaling, and the immune system. (B) Input of the 800 proteins significantly differentially expressed at P < .05 comparing tFL and st-FL samples. The groups contain proteins belonging to the immune system, energy metabolism, cell cycle, cellular growth and activation, and the cytoskeleton. Nodes represent proteins and edges visualize interactions. The different cellular pathways visualized were significantly disturbed in the STRING analysis. Yellow indicates proteins involved in said pathway; blue, protein not involved in the pathway. ATP, adenosine triphosphate; GTPase, guanosine triphosphatase.
Differentially expressed proteins indicate that diagnostic and transformed tumors are molecularly different diseases
Between paired diagnostic st-FL and high-grade transformed tFL samples, the analyses were also restricted to proteins with at least 20% difference in fold changes. Here, 800 proteins were identified as significantly differentially expressed at P < .05 (supplemental Table 5; Figure 5A). Of these, 349 proteins were upregulated in tFL tumors (fold changes, 1.20-6.61) and 451 proteins were downregulated (fold changes, 0.20-0.84). At the P < .01 threshold, 486 proteins were differentially expressed when comparing st-FL and tFL samples. Thus, st-FL and tFL samples showed, as expected, much more abundant proteomic differences and divergency than what was found when comparing diagnostic nt-FL and st-FL samples, which correlated with the accompanying change in histology and cellular composition.
Protein profiles enable complete separation of diagnostic st-FL samples and high-grade tFL samples. (A) Differentially expressed proteins comparing diagnostic st-FL and transformed high-grade tFL samples. The x-axis represents the fold changes as transformed by log2(tFL/st-FL); thus, red dots mark upregulated proteins at P < .05 and a fold change >20%, whereas green dots mark downregulated proteins at P < .05 and a fold change <20%. To allow for better visualization of highly significant data points, the negative logarithm (log10) of the P values are plotted on the y-axis. Horizontal lines mark P values of P < .05 and P < .01, respectively. Vertical dashed lines mark proteins with fold changes of at least 20%. (B) 3D PCA with input of significantly differentially expressed proteins at P < .05 comparing st-FL and tFL samples. (C) 3D PCA with input of differentially expressed proteins at P < .01 comparing st-FL and tFL samples. (D) Hierarchal clustering based on significantly differentially expressed proteins at P < .05 comparing st-FL and tFL samples.
Protein profiles enable complete separation of diagnostic st-FL samples and high-grade tFL samples. (A) Differentially expressed proteins comparing diagnostic st-FL and transformed high-grade tFL samples. The x-axis represents the fold changes as transformed by log2(tFL/st-FL); thus, red dots mark upregulated proteins at P < .05 and a fold change >20%, whereas green dots mark downregulated proteins at P < .05 and a fold change <20%. To allow for better visualization of highly significant data points, the negative logarithm (log10) of the P values are plotted on the y-axis. Horizontal lines mark P values of P < .05 and P < .01, respectively. Vertical dashed lines mark proteins with fold changes of at least 20%. (B) 3D PCA with input of significantly differentially expressed proteins at P < .05 comparing st-FL and tFL samples. (C) 3D PCA with input of differentially expressed proteins at P < .01 comparing st-FL and tFL samples. (D) Hierarchal clustering based on significantly differentially expressed proteins at P < .05 comparing st-FL and tFL samples.
Based on differentially expressed proteins at P < .05 and P < .01, respectively, PCA could fully discriminate between the diagnostic FL and transformed samples, indicating that st-FL and tFL samples are biologically different tumors (Figure 5B-C). Interestingly, the diagnostic st-FL samples showed more focused clustering in the PCAs, whereas tFL samples showed more widespread patterns, which could indicate an increase in disease heterogeneity and tumoral complexity across the transformed tumors. The same observation was true when performing hierarchal clustering, in which the 800 significantly differentially expressed proteins at P < .05 enabled complete separation corresponding to the st-FL and tFL samples (Figure 5D). Taken together, this highlights the marked differences in tumor biology from FL onset to HT.
Disturbed cellular pathways at FL diagnosis and at HT
The STRING analyses were performed on the set of significantly differentially expressed proteins (P < .05). With 798 nodes and 8492 edges, the network had significantly more interactions than expected occur at random (P < .001; Figure 4B). Thus, the analysis of paired st-FL and tFL tumors revealed marked numbers of disturbed pathways, further indicating the striking biological differences present from the time of diagnosis compared with those at transformation. Again, the IPA software algorithms showed similar results to the STRING analysis, with particularly noticeable changes seen influencing cell cycle, cellular energy production, growth, and survival as well as changes in immune response processes, activation, differentiation, and regulation (supplemental Tables 11-13). One must note that in the transformation from FL to tFL, the tumoral cellular composition changes; therefore, differences would be expected solely based on the type of cell analyzed.
Immunohistochemical evaluation of biomarkers validates protein expression patterns and identifies apoptotic deregulation predictive of HT
Interestingly, from the large-scale MS-based study, several proteins involved in apoptotic regulation were identified. Five of those significantly differentially expressed proteins involved in apoptotic signaling were selected for evaluation by immunohistochemical staining. One nt-FL sample was excluded from the analyses because of insufficient lymphoma material being available (n = 33).
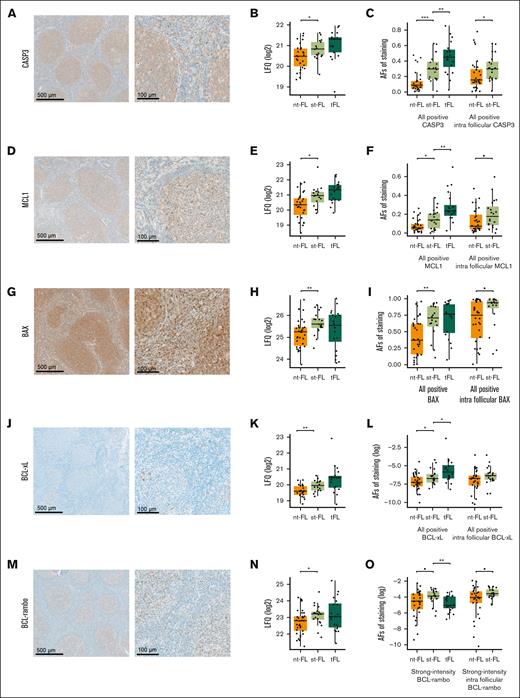
When evaluated by immunohistochemistry, all 5 apoptotic proteins revealed protein expression levels higher in st-FL, which were consistent with the finding in the MS proteomic analyses (Table 2; Figure 6). The expression patterns of CASP3, MCL1, and BAX showed diffuse cytoplasmic staining of both neoplastic and nonneoplastic cells in the TME, whereas expressions of BCL-xL and BCL-rambo were restricted to more specific cellular subsets (Figure 6A,D,G,J,M). For all 5 proteins, most of the positive staining was localized within intrafollicular areas.
Immunohistochemical expression of 5 selected apoptotic biomarkers
| Biomarker . | All, n (%) . | nt-FL, n (%) . | st-FL, n (%) . | P . |
|---|---|---|---|---|
| CASP3 | ||||
| Whole biopsy | <.001 | |||
| High | 27 (50) | 8 (24) | 19 (95) | |
| Low | 27 (50) | 25 (76) | 1 (5) | |
| Intrafollicular | <.001 | |||
| High | 22 (42) | 5 (16) | 17 (85) | |
| Low | 30 (58) | 27 (84) | 3 (15) | |
| MCL1 | ||||
| Whole biopsy | .011 | |||
| High | 19 (36) | 7 (21) | 12 (60) | |
| Low | 34 (64) | 26 (79) | 8 (40) | |
| Intrafollicular | NS | |||
| High | 45 (87) | 26 (81) | 19 (95) | |
| Low | 7 (13) | 6 (19) | 1 (5) | |
| BAX | ||||
| Whole biopsy | .003 | |||
| High | 22 (42) | 8 (24) | 14 (70) | |
| Low | 31 (58) | 25 (76) | 6 (30) | |
| Intrafollicular | .004 | |||
| High | 25 (48) | 10 (31) | 15 (75) | |
| Low | 27 (52) | 22 (69) | 5 (25) | |
| BCL-xL | ||||
| Whole biopsy | .096 | |||
| High | 28 (53) | 14 (42) | 14 (70) | |
| Low | 25 (47) | 19 (58) | 6 (30) | |
| Intrafollicular | .052 | |||
| High | 38 (73) | 20 (63) | 18 (90) | |
| Low | 14 (27) | 12 (38) | 2 (10) | |
| BCL-rambo | ||||
| Whole biopsy | .046 | |||
| High | 33 (61) | 17 (52) | 16 (80) | |
| Low | 20 (38) | 16 (48) | 4 (20) | |
| Intrafollicular | .009 | |||
| High | 43 (83) | 23 (72) | 20 (100) | |
| Low | 9 (17) | 9 (28) | 0 (0) |
| Biomarker . | All, n (%) . | nt-FL, n (%) . | st-FL, n (%) . | P . |
|---|---|---|---|---|
| CASP3 | ||||
| Whole biopsy | <.001 | |||
| High | 27 (50) | 8 (24) | 19 (95) | |
| Low | 27 (50) | 25 (76) | 1 (5) | |
| Intrafollicular | <.001 | |||
| High | 22 (42) | 5 (16) | 17 (85) | |
| Low | 30 (58) | 27 (84) | 3 (15) | |
| MCL1 | ||||
| Whole biopsy | .011 | |||
| High | 19 (36) | 7 (21) | 12 (60) | |
| Low | 34 (64) | 26 (79) | 8 (40) | |
| Intrafollicular | NS | |||
| High | 45 (87) | 26 (81) | 19 (95) | |
| Low | 7 (13) | 6 (19) | 1 (5) | |
| BAX | ||||
| Whole biopsy | .003 | |||
| High | 22 (42) | 8 (24) | 14 (70) | |
| Low | 31 (58) | 25 (76) | 6 (30) | |
| Intrafollicular | .004 | |||
| High | 25 (48) | 10 (31) | 15 (75) | |
| Low | 27 (52) | 22 (69) | 5 (25) | |
| BCL-xL | ||||
| Whole biopsy | .096 | |||
| High | 28 (53) | 14 (42) | 14 (70) | |
| Low | 25 (47) | 19 (58) | 6 (30) | |
| Intrafollicular | .052 | |||
| High | 38 (73) | 20 (63) | 18 (90) | |
| Low | 14 (27) | 12 (38) | 2 (10) | |
| BCL-rambo | ||||
| Whole biopsy | .046 | |||
| High | 33 (61) | 17 (52) | 16 (80) | |
| Low | 20 (38) | 16 (48) | 4 (20) | |
| Intrafollicular | .009 | |||
| High | 43 (83) | 23 (72) | 20 (100) | |
| Low | 9 (17) | 9 (28) | 0 (0) |
Dichotomous high/low biomarker expression was determined as the cutoff value in area fractions from TFS analyses. P values are calculated from a χ2 test or Fisher exact test. P values in bold are significant.
NS, not significant.
Staining patterns and expression levels of 5 selected apoptotic markers. (A) Representative images of immunohistochemical staining patterns of CASP3 expression (left, original magnification ×5; right, original magnification ×20). (B) CASP3 expression levels identified by MS-based proteomics. (C) CASP3 expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (D) Representative images of immunohistochemical staining patterns of MCL1 expression (left, original magnification ×5; right, original magnification ×20). (E) MCL1 expression levels identified by MS-based proteomics. (F) MCL1 expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (G) Representative images of immunohistochemical staining patterns of BAX expression (left, original magnification ×5; right, original magnification ×20). (H) BAX expression levels identified by MS-based proteomics. (I) BAX expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (J) Representative images of immunohistochemical staining patterns of BCL-xL expression (left, original magnification ×5; right, original magnification ×20). (K) BCL-xL expression levels identified by MS-based proteomics. (L) BCL-xL expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (M) Representative images of immunohistochemical staining patterns of BCL-rambo expression (left, original magnification ×5; right, original magnification ×20). (N) BCL-rambo expression levels identified by MS-based proteomics. (O) BCL-rambo expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. ▪P < .1; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. AF, area fraction.
Staining patterns and expression levels of 5 selected apoptotic markers. (A) Representative images of immunohistochemical staining patterns of CASP3 expression (left, original magnification ×5; right, original magnification ×20). (B) CASP3 expression levels identified by MS-based proteomics. (C) CASP3 expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (D) Representative images of immunohistochemical staining patterns of MCL1 expression (left, original magnification ×5; right, original magnification ×20). (E) MCL1 expression levels identified by MS-based proteomics. (F) MCL1 expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (G) Representative images of immunohistochemical staining patterns of BAX expression (left, original magnification ×5; right, original magnification ×20). (H) BAX expression levels identified by MS-based proteomics. (I) BAX expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (J) Representative images of immunohistochemical staining patterns of BCL-xL expression (left, original magnification ×5; right, original magnification ×20). (K) BCL-xL expression levels identified by MS-based proteomics. (L) BCL-xL expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. (M) Representative images of immunohistochemical staining patterns of BCL-rambo expression (left, original magnification ×5; right, original magnification ×20). (N) BCL-rambo expression levels identified by MS-based proteomics. (O) BCL-rambo expression levels identified by immunohistochemical staining in whole biopsy and intrafollicular areas. ▪P < .1; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. AF, area fraction.
Immunohistochemical evaluation showed that at the time of initial diagnosis, samples from patients with st-FL had significantly higher expression of CASP3 (P < .001), MCL1 (P = .015), BAX (P = .003), and BCL-xL (P = .025) than samples from those with nt-FL, whereas BCL-rambo elevation were only trending (P = .057; Figure 6C,F,I,L,O). At the time of HT, expression levels were significantly higher in tFL with regard to CASP3 (P = .008), MCL1 (P = .008), and BCL-xL (P = .044); conversely, expression of BCL-rambo was significantly decreased in tFL compared with st-FL samples (P = .006) (Figure 6O). When quantifying expression levels exclusively localized within intrafollicular areas, a significant difference was retained for CASP3 (P = .030), whereas MCL1 (P = .061), BAX (P = .069), and BCL-rambo (P = .083) retained trending correlations (Figure 6C,F,I,L,O).
Expression levels of all 5 apoptotic markers showed a significantly strong positive correlation to each other (Table 3). In addition, the markers were correlated to either low hemoglobin (hgb) or lymphopenia, suggesting a general hematopoietic cytopenia. Furthermore, CASP3, BCL-xL, and BAX showed tendencies toward a weaker correlation to increasing FL grade (Table 3).
Correlation of biomarkers to each other and to clinicopathological data
| . | CASP3 . | MCL1 . | BAX . | BCL-xL . | BCL-rambo . |
|---|---|---|---|---|---|
| CASP3 | |||||
| MCL1 | ρ = 0.61 P < .001 | ||||
| BAX | ρ = 0.46 P < .001 | ρ = 0.44 P = .001 | |||
| BCL-xL | ρ = 0.43 P = .001 | ρ = 0.40 P = .003 | ρ = 0.34 P = .014 | ||
| BCL-rambo | ρ = 0.31 P = .029 | ρ = 0.40 P = .004 | ρ = 0.47 P < .001 | ρ = 0.48 P < .001 | |
| FL grade | ρ = 0.31 P = .024 | NS | ρ = 0.26 P = .058 | ρ = 0.29 P = .034 | NS |
| hgb | NS | ρ = −0.33 P = .027 | ρ = −0.42 P = .003 | NS | NS |
| Lymphocytes | ρ = −0.39 P = .008 | NS | NS | NS | ρ = −0.28 P = .066 |
| . | CASP3 . | MCL1 . | BAX . | BCL-xL . | BCL-rambo . |
|---|---|---|---|---|---|
| CASP3 | |||||
| MCL1 | ρ = 0.61 P < .001 | ||||
| BAX | ρ = 0.46 P < .001 | ρ = 0.44 P = .001 | |||
| BCL-xL | ρ = 0.43 P = .001 | ρ = 0.40 P = .003 | ρ = 0.34 P = .014 | ||
| BCL-rambo | ρ = 0.31 P = .029 | ρ = 0.40 P = .004 | ρ = 0.47 P < .001 | ρ = 0.48 P < .001 | |
| FL grade | ρ = 0.31 P = .024 | NS | ρ = 0.26 P = .058 | ρ = 0.29 P = .034 | NS |
| hgb | NS | ρ = −0.33 P = .027 | ρ = −0.42 P = .003 | NS | NS |
| Lymphocytes | ρ = −0.39 P = .008 | NS | NS | NS | ρ = −0.28 P = .066 |
P values in bold are significant. NS, not significant.
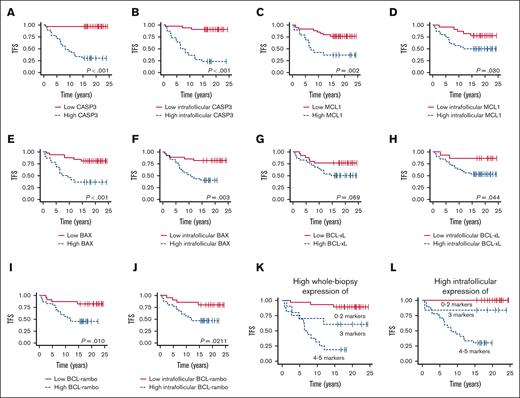
High levels of CASP3, MCL1, BAX, and BCL-rambo expression at the time of initial FL diagnosis were associated with a significantly shorter TFS when analyzing whole tumor tissue biopsy expression (P < .001, P = .002, P < .001, and P = .010, respectively); in contrast, high BCL-xL showed only a trending correlation (P = .069; Figure 7A,C,E,G,I). When analyzing exclusively intrafollicular areas, high expression of all 5 apoptotic markers were associated with significantly inferior TFS (P < .001, P = .030, P = .003, P = .044, and P = .021, respectively; Figure 7B,D,F,H,J).
Outcome according to evaluated apoptotic biomarkers. (A-B) Association between whole biopsy and intrafollicular CASP3 expression and TFS (cutoffs, AF = 0.1423 and AF = 0.3627, respectively). (C-D) Association between whole biopsy and intrafollicular MCL1 expression and TFS (cutoffs, AF = 0.1133 and AF = 0.0264, respectively). (E-F) Association between whole biopsy and intrafollicular BAX expression and TFS (cutoffs, AF = 0.6221 and AF = 0.882, respectively). (G-H) Association between whole biopsy and intrafollicular BCL-xL expression and TFS (cutoffs, AF = 0.0007 and AF = 0.0007, respectively). (I-J) Association between whole biopsy and intrafollicular BCL-rambo expression and TFS (cutoffs, AF = 0.022 and AF = 0.0182, respectively). (K) Association between TFS and high whole tumor coexpression of 0 to 2, 3, and 4 to 5 of the analyzed markers. (L) Association between TFS and high intrafollicular coexpression of 0 to 2, 3, and 4 to 5 of the analyzed markers.
Outcome according to evaluated apoptotic biomarkers. (A-B) Association between whole biopsy and intrafollicular CASP3 expression and TFS (cutoffs, AF = 0.1423 and AF = 0.3627, respectively). (C-D) Association between whole biopsy and intrafollicular MCL1 expression and TFS (cutoffs, AF = 0.1133 and AF = 0.0264, respectively). (E-F) Association between whole biopsy and intrafollicular BAX expression and TFS (cutoffs, AF = 0.6221 and AF = 0.882, respectively). (G-H) Association between whole biopsy and intrafollicular BCL-xL expression and TFS (cutoffs, AF = 0.0007 and AF = 0.0007, respectively). (I-J) Association between whole biopsy and intrafollicular BCL-rambo expression and TFS (cutoffs, AF = 0.022 and AF = 0.0182, respectively). (K) Association between TFS and high whole tumor coexpression of 0 to 2, 3, and 4 to 5 of the analyzed markers. (L) Association between TFS and high intrafollicular coexpression of 0 to 2, 3, and 4 to 5 of the analyzed markers.
Interestingly, combining expression levels of all 5 markers showed increasingly inferior TFS, with increasing numbers of markers with high expression levels. High expressions of 0 to 2, 3, and 4 to 5 markers were associated with a weak, intermediate, and high risk of transformation, respectively, Figure 7K. Notably, this was even more evident when analyzing exclusively intrafollicular areas, with all nt-FL samples having high expression of only ≤2 of the analyzed apoptotic markers (Figure 7L). Thus, our data suggest the possible prediction of transformation based on apoptotic deregulation.
Discussion
Using high-throughput proteomics, we showed that diagnostic FL tumor tissue samples have different protein expression profiles according to the risk of subsequent high-grade transformation. In addition, our study identified candidate proteins for further investigation as possible prognostic and/or predictive biomarkers. Currently, clinical outcomes vary widely among patients with FL, with HT being the leading cause of FL-related death.30 The ability to accurately identify patients at the time of FL diagnosis who are at higher risk of HT would be an important clinical advance.
Notably, we observed disturbances in apoptotic signaling, a pathway already implicated in FL lymphomagenesis as a result of the association of FL with the t(14;18) hallmark translocation.31 The BCL2 protein was also identified in our analyses but without statistically differential expression between the groups. In recent years, research has suggested an association between BCL2 gene mutations and the risk of transformation.32 However, various factors may cause discrepancies between gene expression levels and the final protein product; thus, in this study, we investigated protein expression levels. Indeed, we identified several differentially expressed proteins involved in apoptotic signaling, including CASP3, MCL1, BAX, BCL-xL, and BCL-rambo, which suggests additional apoptotic deregulation in the lymphomas depending on risk of subsequent transformation. Although apoptosis is traditionally considered a barrier to tumorigenesis, it may also instigate proliferation-inducing paracrine effects and contribute to an immunosuppressive TME though phagocytosis by macrophages.33,34 Our results identified upregulation of ubiquitin conjugating enzyme E2 K and proteasome activator subunit 3, which both act in P53 degradation35-40 and P53-regulated apoptotic activator BAX upregulation.41-44 Although TP53 loss is not commonly associated with FL development, it has recurrently been reported in tFL cases.8,45,46 Thus, these results suggest further deregulation mechanisms leading to loss of P53 protein function in addition to commonly known TP53 mutations.
This large-scale proteomic study was performed as a hypothesis-generating investigation, with the aim of identifying novel markers important for HT in FL. In accordance with the results in the MS-based proteomic analysis, immunohistochemical quantification showed elevated expression levels of all 5 investigated apoptotic markers (CASP3, MCL1, BAX, BCL-xL, and BCL-rambo). Furthermore, striking associations were observed between the apoptotic markers and patient prognosis. Importantly, based on expression levels of these markers, we were able to identify patients with markedly inferior TFS. Additionally, all markers showed a strong positive correlation with each other. Thus, our data suggest that in addition to the influence of t(14;18), other factors resulting in apoptotic deregulation may play a role in subsequent risk of transformation in patients with FL. Further studies in larger, independent patient cohorts are warranted to investigate whether these 5, and possibly other, apoptotic proteins are of interest as either prognostic or predictive biomarkers of HT in FL.
Our study also identified other common cancer-associated signaling pathways at play, including a network centered around Rac/Rho guanosine triphosphatase signaling. Interestingly, several proteins involved in these pathways were downregulated in st-FL samples compared with nt-FL samples, including proteins required for regulation of a normal actin cytoskeleton.47 Changes appeared in a β-actin–centered network involving the cytoskeleton. Although often regarded simply as having housekeeping roles, β-actin and the cytoskeleton serve vitally important functions in the cell, including cytokinesis, cell motility, adhesion, migration, and mechanical stability. Moreover, the intracellular actin cytoskeleton assembly is tightly regulated, as many signal transduction systems, such as the Rac/Rho guanosine triphosphatase cycle, use the actin cytoskeleton as a scaffold, and allowing cascading of signal-processing enzymes.47-49 Closely related to this, we found upregulation of many candidates involved in RNA processing and protein metabolism, resulting in an enriched cell cycle progression in st-FL compared with nt-FL samples.
The comparison of protein profiles of st-FL and tFL samples proved to be notably more significant and revealed completely distinct clustering, as expected, because of the differences in disease histologies and cellular composition. To date, the processes driving HT remain largely unknown, although previous studies have suggested increased genomic complexity after HT.11 Our study provides support for this notion at the proteome level, because tFL samples showed much more variable presentations, indicating increased tumoral complexity, and heterogeneity across the high-grade transformed samples.
To the best of our knowledge, studies on MS-based proteomic characterization of FL and transformation of FL are limited. We have previously performed a 2-dimensional (2D)–based proteomics study on a size-limited cohort of patients with FL to investigate transformation.50 Several proteins were identified in both our studies, although some did not reach statistical significance in this study. As we have previously discussed,51 2D gel separation may allow for different proteoforms of the same protein to be analyzed. In some cases, it may be that 1 of these is differentially regulated whereas the other proteoforms are not. The 2D technique focuses on the differentially expressed form. However, the present bottom-up MS-based technique analyzes the combined set of proteoforms, in which the single proteoform that changes may be averaged out. Recently, Duś-Szachniewicz et al13 performed an MS-based study to investigate proteomics of 15 FL samples compared with lymph node control samples. However, the authors provided no information on transformation status or survival, instead highlighting the differences comparing nonneoplastic lymph nodes and FL tumors.13 Among others, CASP3 was shown to be upregulated in FL compared with nonneoplastic lymph nodes, a protein that we also identified as upregulated in st-FL tumors in this study. Using an MS-based approach, Weinkauf et al52 compared FL cell lines with mantle cell lymphoma cell lines, an aggressive B-cell lymphoma entity different from that of DLBCL/tFL. Proteins differentially expressed between the FL and mantle cell lymphoma cell lines were involved in DNA repair, cell cycle control, transcription, and apoptosis, mirroring results from both this study and the study by Duś-Szachniewicz et al.13,52 The authors also identified a specific P53-dependent network of proteins implicated in cell regulation. Also of note, RNA expression data revealed only a modest correlation between RNA and protein levels, emphasizing the relevance of posttranslational regulation in lymphomagenesis.52
We analyzed FL tumor cells in their tissue surroundings, allowing for a better understanding of the biological background behind the neoplastic process.15 Various immune system processes were affected with changes involving B-cell receptor, NF-κB, and PI3K signaling, suggesting variations not only in the B-cell derived lymphoma cells but also differences in the nonneoplastic TME. Especially noteworthy, PAX5, a key B-cell transcription factor, was upregulated in st-FL samples. Our group previously investigated PAX5 expression using immunohistochemistry in samples from the current FL cohort.24 In agreement with the results of this study, our previous evaluation also found PAX5 significantly upregulated in st-FL compared with nt-FL samples.24 With the rapid evolution of modern targeted treatment and personalized medicine, novel therapeutic strategies may not only be aimed directly at tumor cells but also at the components of TME.15,53,54 Thus, an improved understanding of the interplay between neoplastic and nonneoplastic factors may aid the discovery of novel predictive markers of HT. With the presented omics data, we have conducted a comprehensive study of proteins that underlie biological differences among individual FL tumors. These data may predict downstream effects and identify new targets or candidate biomarkers for potential use in future treatment regimens in an era of personalized medicine.
Conclusion
Large-scale proteomics identified important differences in protein profiles in diagnostic FL samples that enabled upfront identification at the time of diagnosis of patients with FL with or without the risk of subsequent transformation. Pathway analyses indicated altered signaling of cellular pathways including apoptosis, cytoskeletal regulation, and cell cycle. Our data identify a novel set of differentially expressed proteins, specifically involved in apoptotic signaling, with the potential to predict at the time of FL diagnosis, the subsequent risk of HT.
Acknowledgments
The authors thank Hanh Pham Hansen, Department of Pathology, Aarhus University Hospital, and Mona Britt Hansen and Ahmed Basim Abduljabar, Department of Biomedicine, Aarhus University, for their excellent technical assistance. The research was funded with grants from Department of Clinical Medicine, Aarhus University, the Karen Elise Jensen Foundation, Merchant Einar Willumsen’s Memorial Foundation, the Danish Lymphoma Group, a donation from Peter and Alice Madsen, Knud and Edith Eriksen’s Memorial Foundation, Eva and Henry Frænkel’s Memorial Foundation, Raimond and Dagmar Ringgård-Bohn’s Foundation, Butcher Max Wørzner and wife Wørzner's Memorial Grant, Master Carpenter Jørgen Holm and wife Elisa F. Hansen’s Memorial Grant, A. P. Møller Foundation for the Advancement of Medical Sciences, Dagmar Marshall’s Foundation, and Farmer of "Ølufgård" Peder Nielsen Kristensens Memorial Foundation. The Orbitrap Fusion Tribrid mass spectrometer was funded by A. P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til almene Formaal.
Authorship
Contribution: M.B.H.E., B.H., and M.L. conceptualized and designed the study; M.B.H.E., K.W., A.J.C., M.D.A., E.F.S., T.E.H., C.M., K.L.L., T.L.P., S.J.H.-D., B.H., and M.L. executed the experiments and statistical analyses; T.L.P. and S.J.H.-D. revised the pathological diagnoses; M.B.H.E. and C.M. acquired clinical data; M.B.H.E. and M.L. wrote the initial draft of the manuscript; and all authors contributed to data interpretation, critically reviewed the manuscript, and approved the final version.
Conflict-of-interest disclosure: B.H. holds shares from Novo Nordisk A/S and Genmab A/S. Novo Nordisk A/S and Genmab A/S had no influence on the study design, analyses, and reporting of results. The remaining authors declare no competing financial interests.
Correspondence: Maja Ludvigsen, Department of Hematology, Aarhus University Hospital, Palle Juul-Jensens Blvd 99, DK-8200 Aarhus N, Denmark; email: majlud@rm.dk; and Bent Honoré, Department of Biomedicine, Aarhus University, The Skou Bldg, Høegh-Guldbergs Gade 10, DK-8000 Aarhus C, Denmark; email: bh@biomed.au.dk.
References
Author notes
Data included in this study are available on reasonable request from the corresponding author, Maja Ludvigsen (majlud@rm.dk).
The full-text version of this article contains a data supplement.