Key Points
B-NHL tumors contained 3 distinct Treg subsets: LAG3+ FOXP3−, resting, and activated Tregs that differed in transcriptome and phenotype.
High abundance of activated Tregs is associated with shorter progression-free survival in follicular lymphoma.
Abstract
Tumor-infiltrating regulatory T cells (Tregs) contribute to an immunosuppressive tumor microenvironment. Despite extensive studies, the prognostic impact of tumor-infiltrating Tregs in B-cell non-Hodgkin lymphomas (B-NHLs) remains unclear. Emerging studies suggest substantial heterogeneity in the phenotypes and suppressive capacities of Tregs, emphasizing the importance of understanding Treg diversity and the need for additional markers to identify highly suppressive Tregs. Here, we applied single-cell RNA sequencing and T-cell receptor sequencing combined with high-dimensional cytometry to decipher the heterogeneity of intratumoral Tregs in diffuse large B-cell lymphoma and follicular lymphoma (FL), compared with that in nonmalignant tonsillar tissue. We identified 3 distinct transcriptional states of Tregs: resting, activated, and unconventional LAG3+FOXP3− Tregs. Activated Tregs were enriched in B-NHL tumors, coexpressed several checkpoint receptors, and had stronger immunosuppressive activity compared with resting Tregs. In FL, activated Tregs were found in closer proximity to CD4+ and CD8+ T cells than other cell types. Furthermore, we used a computational approach to develop unique gene signature matrices, which were used to enumerate each Treg subset in cohorts with bulk gene expression data. In 2 independent FL cohorts, activated Tregs was the major subset, and high abundance was associated with adverse outcome. This study demonstrates that Tregs infiltrating B-NHL tumors are transcriptionally and functionally diverse. Highly immunosuppressive activated Tregs were enriched in tumor tissue but absent in the peripheral blood. Our data suggest that a deeper understanding of Treg heterogeneity in B-NHL could open new paths for rational drug design, facilitating selective targeting to improve antitumor immunity.
Introduction
Regulatory T cells (Tregs) are highly immunosuppressive CD4+ T cells, driven by the transcription factor Forkhead box protein P3 (FOXP3). Although FOXP3 is the master regulator of Tregs, FOXP3-negative Tregs with suppressive function have also been described.2 Tregs act as a double-edged sword by having a protective role in regulating immune homeostasis and a pathological role by inhibiting immune responses in different disease settings such as autoimmunity and cancer.3,4 Aberrantly enriched in the tumor microenvironment (TME), they constitute a potent barrier to effective antitumor immunity by suppressing the activity of effector T cells (Teffs) via a wide range of mechanisms, including production of inhibitory cytokines, perforins, and granzymes; consumption of the critical survival factor interleukin-2 (IL-2); expression of inhibitory checkpoint receptors; and metabolic modulation.5,6 Although Treg biology is well studied and characterized in mice,7-9 human Tregs are currently less well understood. A thorough investigation of their biology is essential before clinical applications can be implemented.
High densities of Tregs infiltrating the TME are associated with poor prognosis in patients with various types of solid cancers,10,11 but the role of Tregs remains controversial in B-cell non-Hodgkin lymphoma (B-NHL). Studies in follicular lymphoma (FL) from the prerituximab era using immunohistochemical analysis reported that higher levels of FOXP3+ cells was associated with better outcome,12,13 whereas other studies found no association14,15 or that higher levels of FOXP3+ cells was associated with poor outcome.16 The conflicting reports might partly be attributed to difficulties in distinguishing Tregs from Teffs, which upon activation transiently express FOXP3 and CD25, thus potentially resulting in the overestimation of the number of intratumoral Tregs, or could be influenced by Treg localization.17,18 A third and less studied possibility is that Tregs found in B-NHL are heterogeneous in terms of their functional state, which may affect treatment responses and clinical outcome.
Tregs were originally considered to be a distinct effector stage of CD4+ T cells with stable suppressive capacity, but emerging evidence indicates that they are phenotypically and functionally diverse, depending on their tissue localization.19,20 Tregs infiltrating human tumors display specific features, indicating that subsets of Tregs may have distinct requirements to function and thrive in a metabolically hostile TME.21-23 Tumor tissue from solid cancers contained highly proliferative and suppressive Tregs, characterized by high expression of checkpoint receptors, such as cytotoxic T-lymphocyte-associated protein 4 (CTLA4), programmed cell death protein 1 (PD-1), T-cell immunoreceptor with Ig and ITIM domains (TIGIT), lymphocyte-activating gene 3 (LAG3), and T-cell immunoglobulin and mucin-domain containing-3 (TIM-3).22,24,25 It is generally accepted that the clinical benefits after immune checkpoint therapy are due to restoration of Teff–mediated antitumor activity, but increasing evidence indicates that Tregs also play a role. The therapeutic effect of the anti-CTLA4 monoclonal antibody ipilimumab has partly been attributed to the depletion of CTLA4+ Tregs in the TME via antibody-dependent cellular cytotoxicity.26-28 Similarly, depletion of Tregs was an important mechanism of action for agonistic antibodies targeting OX40,29 and the suppression of Tregs was observed when OX40 and a TLR9 ligand, administered locally, could cure multiple types of systemic cancer in syngeneic mouse models.30 Tumor-infiltrating Tregs need special attention in relation to immune checkpoint blockade because PD-1 blockade can enhance the suppressive function and proliferation of Tregs and is associated with poorer response and hyperprogressive disease.31,32 Identification of tumor-resident Tregs with high immunosuppressive activity could have important implications for various forms of immunotherapy, including checkpoint blockade, bispecific T-cell engagers, and chimeric antigen receptor (CAR) T-cell therapy.
Here, by investigating intratumoral Tregs on the phenotypic, functional, and transcriptional levels, we identified a subset of activated Tregs that is highly enriched in B-NHL tumors. These activated Tregs are characterized by high coexpression of checkpoint receptors, high immunosuppressive activity, and a distinct transcriptional signature, as assessed by single-cell RNA sequencing (scRNA-seq). Because intratumoral Tregs differ from peripheral blood Tregs, they represent an attractive target for cancer immunotherapy.
Material and methods
Human samples
All samples were obtained with informed written consent in accordance with the Declaration of Helsinki and with approval from the Regional Committee for Medical and Health Research Ethics. Lymph node (LN) specimens were obtained before treatment from patients with FL (n = 20), diffuse large B-cell lymphoma (DLBCL; n = 17), or mantle cell lymphoma (MCL; n = 10) at Oslo University Hospital, Norway. Tonsils were obtained from patients (n = 12) undergoing tonsillectomy (Agroklinikken, Asker). Samples were mechanically dissociated and cryopreserved. Peripheral blood was collected from patients with FL (n = 9) and from anonymous, healthy donors (n = 5) at Oslo University Hospital, processed to obtain peripheral blood mononuclear cells (PBMCs) by Ficoll gradient centrifugation, and cryopreserved. For clinical characteristics and samples overview, see supplemental Tables 1 and 2.
scRNA-seq
Single-cell suspensions were stained with Alexa750, fluorochrome-coupled–, cellular indexing of transcriptomes and epitopes with sequencing (CITE-seq)–, and TotalSeq-C hashtag antibodies (supplemental Table 3). Live CD4+ T cells were sorted on a fluorescence-activated cell sorter (FACS) Aria (BD Biosciences) and resuspended in phosphate-buffer saline (PBS) with 0.05% bovine serum albumin (BSA) to yield 1000 cells per μL.
Using 10x Genomics, sequencing libraries were generated for gene expression (Single Cell 5′ Library kit), T-cell receptor (TCR) (Variable diversity joining Enrichment kit for Human T Cell), and antibody analysis (5′ Feature Barcode Library Kit), according to the manufacturer’s protocols. All libraries were sequenced together on an Illumina NextSeq500 at 2 × 150 base pairs, with an average depth of ∼45 000 read pairs per cell. Cell Ranger (version 3.0.2, 10x Genomics) was used to demultiplex the raw base-call files and convert them into Fastq files that were aligned to the reference genome (hg38). Seurat Package (version 4) was used for quality control and normalization on the count matrices. Counts were adjusted for cell-specific sampling (normalized) using the scTransform function. After stringent filtering, transcriptomes were obtained from a total of 18 771 cells, of which 6230 were from healthy donor tonsils (n = 3), 5712 from patients with FL (n = 3), and 6829 from patients with DLBCL (n = 3).
scRNA-seq bioinformatics
All scRNA-seq data were processed and analyzed using Seurat (version 4).33 FindAllMarkers function was applied to identify differential gene expression, and the results used for gene set enrichment analysis on Human Molecular Signatures Database (MSigDB) Hallmark and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways,34,35 by rapid integration of term annotation and network resources (RITAN) and enrichR.
Cell Ranger (version 3.0.2) for variable diversity joining sequence assembly was applied for TCR reconstruction and paired TCR clonotype calling. From each sample, 1000 random cells with both gene expression and single TCRα/TCRβ genes were retained for TCR analysis using scRepertoire (version 1.7.2).
Building signature matrices and deconvolution of bulk gene expression with CIBERSORTx
Signature matrices from our scRNA-seq data were built by applying CIBERSORTx (https://cibersortx.stanford.edu/), using 2 samples as reference profiles to build the signature matrices, with the remaining 7 samples used for the validation of performance.36-38
We applied the Treg-signature matrices to the discovery bulk FL data set, Steen et al39 and to the British Columbia Cancer Agency (BCCA) FL data set40 using CIBERSORTx S-mode batch correction, and z normalized the resulting cell type abundance. The discovery FL cohort39 was stratified into 2 groups of high and low normalized activated Tregs abundance across a range of percentiles, from 5% to 95%, with intervals of 10%. The activated Treg abundance threshold at the 85th percentile was identified to best separate from the 2 groups in a Kaplan-Meier analysis of progression-free survival. The same threshold (activated Tregs ≥ 0.7511) was subsequently applied to stratify patients in the BCCA FL cohort.40
Phenotypical characterization and spatial analysis
Single-cell suspensions from PBMC (n = 5) and tonsils (n = 10) from healthy donors and patients with FL (n = 15), DLBCL (n = 17), and MCL (n = 10) were stained with antibodies for mass cytometry or fluorescence flow cytometry (supplemental Tables 3 and 4) and acquired by cytometry by time of flight (Standard BioTools) or a multilazer flow cytometer (LSRII, BD Biosciences). Data were analyzed using Enterprise Cytobank (https://cellmass.cytobank.org). Imaging mass cytometry (IMC) was performed on 4-μm tissue section of formalin-fixed, paraffin-embedded FL samples (n = 4). After antigen retrieval, antibody staining (supplemental Table 5) at 4°C, overnight. Tissue sections were ablated in Hyperion Imaging Systems (Standard BioTools).
Treg suppression assay
Proliferation assay was performed as described41 using FAC-sorted Tregs as suppressor cells and autologous CellTrace Violet–labeled Teffs as responder cells from FL (n = 9) and tonsillar (n = 6) samples.
Data scripts
See supplemental Methods for detailed protocols.
Results
Single-cell atlas of CD4+ T cells reveals 3 distinct Treg subsets in lymphoma
For in-depth profiling of intratumoral Tregs in B-NHL, we performed 5′ scRNA-seq paired with CITE-seq and single-cell variable diversity joining sequencing on CD4+ T cells from malignant LNs from patients with FL or DLBCL, and tonsils from healthy donors (Figure 1A). After stringent quality control (supplemental Figure 1A-B), we retained the transcriptomes of 18 771 CD4+ T cells (n = 9; mean of 2086 cells per donor). We performed unsupervised clustering using a weighted nearest neighbor framework33 on the scRNA-seq and CITE-seq data, which identified 13 different CD4+ T-cell clusters (Figure 1B; supplemental Figure 2). We projected the sample type origin of each cell on a uniform manifold approximation and projection (UMAP) and observed that all clusters were populated by cells from several patients (Figure 1C; supplemental Figure 1C-D). Based on cluster genes, the 13 clusters were annotated as 1 naive cluster (KLF2, CCR7, and LEF1), 3 memory clusters (CD69 and SELL), 1 GZM+ cluster (GZMK/A, NKG7, and CST7), 2 follicular helper cell (Tfh) clusters (PDCD1, CXCR5, IL21, and TOX2), 1 T helper cell cluster (CXCR4 and KLF6), 2 conventional Treg clusters (FOXP3, IL2RA, and CTLA4), and a third cluster of unconventional Tregs annotated as LAG3+ Tregs, characterized by high expression of LAG3, CTLA4, and KLRB1 but lacked expression of FOXP3 (Figure 1D; supplemental Figure 3). As expected, DLBCL contributed less to Tfh clusters than FL and tonsils, and tonsils contributed less to the GZM+ cluster and the 2 conventional Treg populations, activated and resting Tregs (Figure 1C). Activated Tregs were characterized by high gene expression of FOXP3, IL32, CTLA4, and TNFRSF18 and high protein levels of TIGIT, OX40, and CD25 but lacked the expression of CD127, as assessed by CITE-seq (Figure 1D-E; supplemental Figure 4A). Trajectory analysis suggested that activated Tregs are closest to, but more differentiated than resting Tregs (supplemental Figure 4B). Next, we investigated the cellular abundance of the Treg subsets across patient samples and observed an enrichment of activated Tregs in FL (7.9%-21.6%) and DLBCL (6.3%-14.6%), in contrast to healthy donor tonsils (1.3%-3.7%; Figure 1F; supplemental Figure 4C-D). Although LAG3+ Tregs dominated in tonsillar samples (7.3%-12.6%), this subset had low abundance in FL (1.2%-7.2%) and DLBCL (0.04%-0.1%). Collectively, these single-cell transcriptome data revealed 3 subsets of Tregs and pointed to the enrichment of activated Tregs in lymphoma tumors, whereas LAG3+ Tregs were predominant in tonsillar samples.
Transcriptional landscape of CD4+ T cells in B-NHL and healthy donor tonsils. (A) Schematic representation of the workflow of scRNA-seq, TCR single-cell variable diversity joining sequencing (scVDJ-seq), and CITE-seq of sorted CD4+ T cells. (B) Single-cell data derived from sorted CD4+ T-cell populations from healthy donor tonsils (n = 3), FL (n = 3), and DLBCL (n = 3) projected onto uniform manifold approximation and projection (UMAP) by combining scRNA-seq and CITE-seq using weighted nearest neighbor (Wnn) method, provided 13 distinct clusters based on gene and protein expression differences for 18 771 cells passing the quality control. The clusters were annotated based on a combination of gene and protein expression as naive (KLF2, CCR7, LEF1, and protein expression of CD45RA), 3 memory clusters (CD69, SELL, and lack of CD45RA protein expression), GZM+ CD4+ T cells (GZMK/A, NKG7, and CST7), 2 T follicular helper (Tfh) clusters (PDCD1, CXCR5, IL21, TOX2, and protein expression of PD-1, CXCR5), a T helper cell cluster (CXCR4 and KLF6), 2 clusters of Tregs (FOXP3, IL2RA, CTLA4, and protein expression of CD25 but lack of CD127), and 1 cluster of unconventional FOXP3−LAG3+ Tregs (LAG3, CTLA4, IL10, and lack of CD127 protein). (C) UMAPs of the single-cell data shown in panel B, divided based on tissue origin and color coded based on patient sample. (D) Dot plots showing average expression of top 5 differentially expressed genes (DEGs) for each cluster. (E) Expression of selected genes (purple) and surface protein expression (green) overlaid onto the Wnn UMAP coordinates from panel B. (F) Pie charts showing the cellular abundance of each cell cluster per patient sample.
Transcriptional landscape of CD4+ T cells in B-NHL and healthy donor tonsils. (A) Schematic representation of the workflow of scRNA-seq, TCR single-cell variable diversity joining sequencing (scVDJ-seq), and CITE-seq of sorted CD4+ T cells. (B) Single-cell data derived from sorted CD4+ T-cell populations from healthy donor tonsils (n = 3), FL (n = 3), and DLBCL (n = 3) projected onto uniform manifold approximation and projection (UMAP) by combining scRNA-seq and CITE-seq using weighted nearest neighbor (Wnn) method, provided 13 distinct clusters based on gene and protein expression differences for 18 771 cells passing the quality control. The clusters were annotated based on a combination of gene and protein expression as naive (KLF2, CCR7, LEF1, and protein expression of CD45RA), 3 memory clusters (CD69, SELL, and lack of CD45RA protein expression), GZM+ CD4+ T cells (GZMK/A, NKG7, and CST7), 2 T follicular helper (Tfh) clusters (PDCD1, CXCR5, IL21, TOX2, and protein expression of PD-1, CXCR5), a T helper cell cluster (CXCR4 and KLF6), 2 clusters of Tregs (FOXP3, IL2RA, CTLA4, and protein expression of CD25 but lack of CD127), and 1 cluster of unconventional FOXP3−LAG3+ Tregs (LAG3, CTLA4, IL10, and lack of CD127 protein). (C) UMAPs of the single-cell data shown in panel B, divided based on tissue origin and color coded based on patient sample. (D) Dot plots showing average expression of top 5 differentially expressed genes (DEGs) for each cluster. (E) Expression of selected genes (purple) and surface protein expression (green) overlaid onto the Wnn UMAP coordinates from panel B. (F) Pie charts showing the cellular abundance of each cell cluster per patient sample.
Treg subsets share clonotypes in FL
Antigen and environmental cues can guide Treg differentiation to an activated phenotype and increased suppressive activity. We investigated the TCR repertoire and found little clonal expansion across the CD4+ T-cell clusters, except for GZM+ T cells that showed the highest level of expansion (Figure 2A-B; supplemental Figure 5A-B). Treg clusters also showed limited expansion and similar diversity score as measured by the Shannon diversity index (Figure 2B). Although most Tregs had unique clonotypes, we observed medium expansion of clonotypes (3-6 cells sharing the same clonotype) in activated Tregs in DLBCL (Figure 2C). Next, we measured the degree to which the frequency distribution of CDR3 lengths deviated from normality as an indication of clonal expansion.42 Concordant with the clonal expansion data, activated Treg clusters in DLBCL and FL demonstrated significant deviation for CDR3 length, as calculated by the Shapiro-Wilk test43 (P < .001), but activated Tregs in DLBCL had the lowest W value, of 0.971, and a highest skew, measured by the D’Agostino test (Table 1), consistent with having the highest clonal expansion among Tregs (Figure 2D; supplemental Figure 5C).
Characterization of TCR diversity and clonotype expansion by scVDJ-seq. (A) Clonal expansion mapped onto the UMAP from Figure 1B. (B) Shannon diversity index estimated from TCR sequencing results for 45 random cells per cluster. Values are shown for each cluster identified in Figure 1B. (C) Histograms showing the percentages of Treg subsets with expanded clonotypes within the entire Treg compartment and across samples of tonsils (n = 3), FL (n = 3), and DLBCL (n = 3). Expanded clonotypes were categorized as small expansion (1 < X ≤ 3) and medium expansion (3 < X ≤ 6). (D) Distribution of CDR3 length for each Treg cluster as per the tissue origin. The mean CDR3 lengths for activated Tregs (actTregs), resting Tregs (restTregs), and LAG3+ Tregs were 28.1 amino acids (aas), 28.3 aas, and 27.9 aas, respectively in FL; 28.0 aas, 27.7 aas, and 28.3 aas in DLBCL; and 28.3 aas, 28.0 aas, and 28.1 aas in tonsils. (E) Venn diagrams showing the number of clonotypes within and shared between Treg subsets. For scaling purposes, numbers are shown for the Tregs found among 1000 random CD4+ T cells with gene expression and single TCRα/TCRβ gene.
Characterization of TCR diversity and clonotype expansion by scVDJ-seq. (A) Clonal expansion mapped onto the UMAP from Figure 1B. (B) Shannon diversity index estimated from TCR sequencing results for 45 random cells per cluster. Values are shown for each cluster identified in Figure 1B. (C) Histograms showing the percentages of Treg subsets with expanded clonotypes within the entire Treg compartment and across samples of tonsils (n = 3), FL (n = 3), and DLBCL (n = 3). Expanded clonotypes were categorized as small expansion (1 < X ≤ 3) and medium expansion (3 < X ≤ 6). (D) Distribution of CDR3 length for each Treg cluster as per the tissue origin. The mean CDR3 lengths for activated Tregs (actTregs), resting Tregs (restTregs), and LAG3+ Tregs were 28.1 amino acids (aas), 28.3 aas, and 27.9 aas, respectively in FL; 28.0 aas, 27.7 aas, and 28.3 aas in DLBCL; and 28.3 aas, 28.0 aas, and 28.1 aas in tonsils. (E) Venn diagrams showing the number of clonotypes within and shared between Treg subsets. For scaling purposes, numbers are shown for the Tregs found among 1000 random CD4+ T cells with gene expression and single TCRα/TCRβ gene.
Shapiro-Wilk normality test and D'Agostino skewness test for CDR3 length distribution across Treg subsets
| . | Shapiro-Wilk normality test . | D'Agostino skewness test . | ||
|---|---|---|---|---|
| W value . | P value . | Skew . | P value . | |
| FL | ||||
| actTregs | 0.980 | <.001 | 0.235 | .015 |
| restTregs | 0.977 | <.001 | 0.067 | .509 |
| LAG3+ Tregs | 0.972 | <.01 | −0.234 | .256 |
| DLBCL | ||||
| actTregs | 0.971 | <.001 | 0.311 | .009 |
| restTregs | 0.976 | .049 | 0.149 | .508 |
| Tonsils | ||||
| actTregs | 0.974 | .107 | −0.039 | .878 |
| restTregs | 0.976 | .186 | −0.006 | .981 |
| LAG3+ Tregs | 0.980 | <.001 | 0.079 | .493 |
| . | Shapiro-Wilk normality test . | D'Agostino skewness test . | ||
|---|---|---|---|---|
| W value . | P value . | Skew . | P value . | |
| FL | ||||
| actTregs | 0.980 | <.001 | 0.235 | .015 |
| restTregs | 0.977 | <.001 | 0.067 | .509 |
| LAG3+ Tregs | 0.972 | <.01 | −0.234 | .256 |
| DLBCL | ||||
| actTregs | 0.971 | <.001 | 0.311 | .009 |
| restTregs | 0.976 | .049 | 0.149 | .508 |
| Tonsils | ||||
| actTregs | 0.974 | .107 | −0.039 | .878 |
| restTregs | 0.976 | .186 | −0.006 | .981 |
| LAG3+ Tregs | 0.980 | <.001 | 0.079 | .493 |
actTregs, activated Tregs; restTregs, resting Tregs.
We observed that several TCR clonotypes were shared between the activated and resting Tregs in 2 of 3 FL samples, whereas only 1 clonotype was shared between these Treg subsets in DLBCL (Figure 2E). In each case, the shared clonotypes were not expanded within 1 cluster. In contrast, no clonotypes were shared between Treg subsets in the tonsillar samples (Figure 2E). These data suggest that although Tregs are enriched in B-NHL, the clonal expansion in the TME is limited and mostly restricted to the same Treg subset. However, for tumor-resident Tregs in FL, activated and resting Tregs shared clonotypes, suggesting that intratumoral Tregs are dynamic and display more plasticity.
Intratumoral Treg subsets have distinct transcriptional signatures
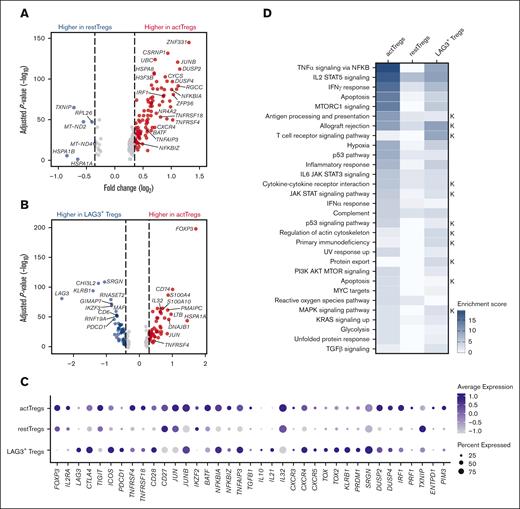
To further characterize the transcriptional signatures distinguishing the 3 Treg subsets, we applied single-cell differential gene expression profiling. The majority of genes that were differentially expressed between activated and resting Tregs were upregulated in activated Tregs, suggesting a higher transcriptional activity in this subset (Figure 3A). Activated Tregs had higher expression levels of checkpoint receptors (TNFRSF4 and TNFRSF18) and genes involved in immune response activation (DUSP2 and DUSP4)44 and the NF-κB pathway (NFKBIA, TNFAIP3, and NFKBIZ) (Figure 3A; supplemental Table 6). Resting Tregs had upregulated stress response genes (HSPA1A and HSPA1B). Comparing activated Tregs with LAG3+ Tregs, activated Tregs had higher expression of FOXP3, major histocompatibility complex class II–associated invariant chain (CD74), and proinflammatory cytokines (IL32 and LTB), whereas LAG3+ Tregs had higher expression of LAG3 and immunosuppressive signature genes (KLRB1 and IL10) (Figure 3B; supplemental Table 7). Activated Tregs had the highest expression of TIGIT, TNFRSF4, and TNFRSF18, the transcription factors JUN and IRF1, and the immunosuppressive cytokines IL32 and TGFB1 (Figure 3C). We applied single-cell gene set enrichment analysis and identified tumor necrosis factor α signaling via NF-κB, IL-2 receptor subunit α–dependent activation of the transcription factor STAT5, and interferon-γ signaling response as top pathways in activated Tregs compared with those in resting and LAG3+ Tregs (Figure 3D; supplemental Figure 6). These pathways have an essential role in Treg homeostasis and suppressive function.45-47
Characterization of Treg subsets by scRNA-seq. Volcano plots based on the scRNA-seq data from healthy donor tonsils (n = 3), FL (n = 3), and DLBCL (n = 3), showing key significantly DEGs between (A) activated and resting Tregs and (B) activated and LAG3+ Tregs. Each dot indicates 1 gene; red dots represent a log2 fold change (log2FC) of >0.35, and blue dots depict a log2FC < −0.35. Significant genes were selected with adjusted P value < .05. Full list of DEGs is shown in supplemental Tables 6 and 7. (C) Dot plot of selected genes from scRNA-seq data, showing average gene expression of checkpoint receptors, transcription factors, cytokines, and DEGs across the 3 distinct cellular subsets of Tregs. (D) Heat map of gene set enrichment analysis (GSEA) highlighting highly enriched pathways among the 3 Treg clusters based of Hallmark and KEGG (K) libraries, shown as enrichment score.
Characterization of Treg subsets by scRNA-seq. Volcano plots based on the scRNA-seq data from healthy donor tonsils (n = 3), FL (n = 3), and DLBCL (n = 3), showing key significantly DEGs between (A) activated and resting Tregs and (B) activated and LAG3+ Tregs. Each dot indicates 1 gene; red dots represent a log2 fold change (log2FC) of >0.35, and blue dots depict a log2FC < −0.35. Significant genes were selected with adjusted P value < .05. Full list of DEGs is shown in supplemental Tables 6 and 7. (C) Dot plot of selected genes from scRNA-seq data, showing average gene expression of checkpoint receptors, transcription factors, cytokines, and DEGs across the 3 distinct cellular subsets of Tregs. (D) Heat map of gene set enrichment analysis (GSEA) highlighting highly enriched pathways among the 3 Treg clusters based of Hallmark and KEGG (K) libraries, shown as enrichment score.
Intratumoral Tregs are skewed toward activated Tregs with high immunosuppressive activity
Next, we investigated whether the transcriptional Treg clusters could be identified as distinct Treg phenotypes, which would facilitate cell sorting and functional assessment. Based on the scRNA-seq and CITE-seq data, we developed an antibody panel for mass cytometry enabling identification of T-cell subsets and expression of 18 different checkpoint receptors (supplemental Table 4). We focused on CD4+CD25+FOXP3+ Tregs because these were dominant in the malignant tissues. Unsupervised clustering of FOXP3+CD25+CD4+ T cells using FlowSOM demonstrated that 2 of 4 metaclusters, metacluster 1 and 2, were completely dominant (Figure 4A). Based on low/lack of checkpoint receptor expression, metacluster 1 corresponded to the resting Treg cluster, whereas metacluster 2 corresponded to the activated Treg cluster with high expression of several checkpoint receptors, including CTLA4, TIGIT, PD-1, ICOS, and OX40, and activation markers, including CD45RO and CD25 (Figure 4A-B; supplemental Figure 7A). This is in line with the CITE-seq data, showing that the 3 Treg clusters were separated based on protein expression of TIGIT, ICOS, OX40, PD-1, and CD25 (supplemental Figures 2B and 4A). The abundance of activated Tregs estimated by scRNA-seq was highly concordant with the mass cytometry data (supplemental Figure 7B).
Intratumoral Tregs are phenotypically and functionally heterogeneous. (A) The expression of 18 checkpoint receptors on CD4+FOXP3+CD25+CD127− Tregs from healthy donor PBMCs (n = 3) and tonsils (n = 3), FL (n = 3), and DLBCL (n = 2) were assessed by t-distributed stochastic neighbor embedding (tSNE) analysis and FlowSOM clustering of mass cytometry data (supplemental Table 1). (Top) Population analysis of Tregs colored based on FlowSOM metaclusters. (Bottom) Density plots of checkpoint receptor protein expression on concatenated Tregs. (B) Heat map of checkpoint receptor expression in activated Tregs (metacluster 2) and resting Tregs (metacluster 1) from the same samples as in panel A. The heat map shows arcsinh ratio of median fluorescence intensity (MFI) normalized to column minimum. (C) Spearman correlation of Treg subset distribution assessed by flow cytometry (5-marker tSNE) with the distribution assessed by mass cytometry (18-marker tSNE). (D) Frequency of activated Tregs among CD4+ T cells assessed by flow cytometry in PBMCs (n = 5) and tonsils (n = 7) from healthy donors, and in FL (n = 15), DLBCL (n = 16), and MCL (n = 10). Horizontal line represents median frequency. (E) Frequency of the activated Treg subset among CD4+ T cells in paired samples, PBMCs, and LNs, from patients with FL (n = 9). (F) Representative flow cytometry histogram of CellTrace Violet dilution in effector CD4+ or CD8+ T cells when stimulated with T-cell-expansion beads (stim. Teff) and cocultured in presence of either actTregs or restTregs purified from an FL sample (left), or calculated as percent suppression of stim. Teff proliferation when cocultured in the presence vs absence of Tregs, (n = 9). (G) (Left) A representative experiment showing TGF-β expression for activated and resting Tregs in the presence or absence of different stimuli. (Right) Summary of TGF-β median fluorescence intensity for unstimulated, activated, and resting Tregs from FL (n = 4) and tonsillar (n = 5) samples. Statistical differences were calculated using Kruskal-Wallis with Dunn multiple comparison test for panel D and Wilcoxon for panels E-G. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Intratumoral Tregs are phenotypically and functionally heterogeneous. (A) The expression of 18 checkpoint receptors on CD4+FOXP3+CD25+CD127− Tregs from healthy donor PBMCs (n = 3) and tonsils (n = 3), FL (n = 3), and DLBCL (n = 2) were assessed by t-distributed stochastic neighbor embedding (tSNE) analysis and FlowSOM clustering of mass cytometry data (supplemental Table 1). (Top) Population analysis of Tregs colored based on FlowSOM metaclusters. (Bottom) Density plots of checkpoint receptor protein expression on concatenated Tregs. (B) Heat map of checkpoint receptor expression in activated Tregs (metacluster 2) and resting Tregs (metacluster 1) from the same samples as in panel A. The heat map shows arcsinh ratio of median fluorescence intensity (MFI) normalized to column minimum. (C) Spearman correlation of Treg subset distribution assessed by flow cytometry (5-marker tSNE) with the distribution assessed by mass cytometry (18-marker tSNE). (D) Frequency of activated Tregs among CD4+ T cells assessed by flow cytometry in PBMCs (n = 5) and tonsils (n = 7) from healthy donors, and in FL (n = 15), DLBCL (n = 16), and MCL (n = 10). Horizontal line represents median frequency. (E) Frequency of the activated Treg subset among CD4+ T cells in paired samples, PBMCs, and LNs, from patients with FL (n = 9). (F) Representative flow cytometry histogram of CellTrace Violet dilution in effector CD4+ or CD8+ T cells when stimulated with T-cell-expansion beads (stim. Teff) and cocultured in presence of either actTregs or restTregs purified from an FL sample (left), or calculated as percent suppression of stim. Teff proliferation when cocultured in the presence vs absence of Tregs, (n = 9). (G) (Left) A representative experiment showing TGF-β expression for activated and resting Tregs in the presence or absence of different stimuli. (Right) Summary of TGF-β median fluorescence intensity for unstimulated, activated, and resting Tregs from FL (n = 4) and tonsillar (n = 5) samples. Statistical differences were calculated using Kruskal-Wallis with Dunn multiple comparison test for panel D and Wilcoxon for panels E-G. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
We investigated the frequency of Treg subsets in a larger cohort of patients with B-NHL by flow cytometry, using a smaller panel of checkpoint receptors. For samples that were phenotyped by fluorescence flow and mass cytometry, there was a high concordance between cellular abundance of activated Tregs as identified by either method, validating the use of a smaller panel to distinguish Treg subsets (Spearman correlation, r = 0.95; Figure 4C; supplemental Figure 7C). Enumeration by flow cytometry demonstrated that activated Tregs were significantly enriched in lymphoma tumors with a median frequency of 14.6% (1.3%-27.9%) of total CD4+ T cells in FL, 10.2% (2.1%-84.2%) in DLBCL, and 7.4% (4.9%-24.9%) in MCL, as compared with 0.3% (0.2%-0.7%) and 3.5% (2.1%-5.2%) in healthy donor PBMCs and tonsils, respectively (Figure 4D). In contrast, resting Tregs were present at low frequencies in B-NHL tumors (0.5%-16.3%), similar to healthy donor PBMCs (2.7%-4.5%) and tonsils (0.9%-2.7%), demonstrating that an expansion of activated Tregs accounts for the increased frequency of Tregs in lymphoma (supplemental Figure 7D-E). Analysis of paired PBMCs and malignant LNs from patients with FL further demonstrated a significantly higher frequency of activated Tregs in tumors compared with that in the peripheral blood (Figure 4E). Together, these results clearly demonstrate that activated Tregs is the main Treg subset in B-NHL tumors, in contrast to a very low frequency of this subset in tonsils and PBMCs.
To investigate whether the phenotypic diversity among intratumoral Tregs translates into functional diversity, we tested the capacity of Tregs to inhibit proliferation of autologous intratumoral CD4+ and CD8+ Teffs. Teffs were subjected to FACS, labeled with Cell Trace Violet before coculturing, and then activated by T-cell-activation beads for 4 days in the presence of purified autologous resting or activated Tregs (supplemental Figure 8A-C). These experiments demonstrated that resting and activated Tregs from FL had immunosuppressive activity on autologous Teffs (Figure 4F). Strikingly, activated Tregs were significantly more potent than resting Tregs in suppressing the proliferation of CD4+ and CD8+ Teffs (Figure 4F). Similar results were observed for healthy donor tonsils (supplemental Figure 8D). To further characterize the immunosuppressive capacity of Treg subsets, we investigated cytokine production and found that activated Tregs had higher levels of transforming growth factor β and IL-10 than resting Tregs (Figure 4G; supplemental Figure 8E-F). These results indicate that the phenotypical diversity of Tregs is associated with differences in their functional state.
Activated Tregs are found in close proximity to T cells in FL
The TME in FL is characterized by numerous malignant follicles containing mainly lymphoma cells and interfollicular zones enriched for T cells. We investigated the localization of activated Tregs in FL and their spatial relationship with neighboring cells by IMC. Based on our phenotypic characterization of Tregs and the availability of well-working antibodies for IMC, we defined activated Tregs as CD20−CD3+CD4+FOXP3+PD-1+TIGIT+. Images revealed the presence of activated Tregs in the malignant follicle and in the T-cell–rich zone (Figure 5A; supplemental Figure 9). To investigate cell–cell interactions based on spatial proximity, we calculated the distance from all tumor and immune cells to the nearest activated Treg, independent of zones (Figure 5B). The median distance to the nearest activated Treg was lower for CD4+ and CD8+ T cells as well as macrophages than for tumor cells, Tfh, dendritic cells, and GZM+ cells (Figure 5B-D; supplemental Table 8). This indicates that although activated Tregs were distributed throughout the TME in FL, they preferentially interact with other T cells, supporting a key role in immunosuppression of Teffs.
Spatial single-cell analysis of FL tissues. Imaging mass cytometry was performed on tissue section from formalin-fixed paraffin-embedded FL samples (n = 4). (A) Shown is 1 representative image with FOXP3 (green, Tregs), CD4 (pink), CD20 (blue; tumor cells), and CD21 (yellow; follicular dendritic cells) delimiting the follicles and T-cell zone. Scale bar, 100 μm. (B) Scatter plots depicting the median distance of different cell populations to the nearest actTreg, localized either in the T-cell–rich zone or in the intrafollicular area. Statistical differences were calculated using Kruskal-Wallis test and Dunn multiple comparisons (supplemental Table 8). ∗∗∗P < .001 and ∗∗∗∗P < .0001. (C) actTregs can colocalize with CD8 T cells. Shown is a selected area from the T-cell zone of panel A, with FOXP3 (green), TIGIT (pink), and CD8 (blue). Arrow heads point at FOXP3+TIGIT+ actTregs in proximity to CD8+ T cells. (D) actTregs can colocalize with dendritic cells (DCs) and macrophages. Shown is 1 representative area from the image in panel A, with FOXP3 (green), CD11c (pink, DCs), CD68 (blue, macrophages), and CD21 (white). Arrow heads point at Tregs in proximity to DCs, whereas arrows point at Tregs in proximity to macrophages.
Spatial single-cell analysis of FL tissues. Imaging mass cytometry was performed on tissue section from formalin-fixed paraffin-embedded FL samples (n = 4). (A) Shown is 1 representative image with FOXP3 (green, Tregs), CD4 (pink), CD20 (blue; tumor cells), and CD21 (yellow; follicular dendritic cells) delimiting the follicles and T-cell zone. Scale bar, 100 μm. (B) Scatter plots depicting the median distance of different cell populations to the nearest actTreg, localized either in the T-cell–rich zone or in the intrafollicular area. Statistical differences were calculated using Kruskal-Wallis test and Dunn multiple comparisons (supplemental Table 8). ∗∗∗P < .001 and ∗∗∗∗P < .0001. (C) actTregs can colocalize with CD8 T cells. Shown is a selected area from the T-cell zone of panel A, with FOXP3 (green), TIGIT (pink), and CD8 (blue). Arrow heads point at FOXP3+TIGIT+ actTregs in proximity to CD8+ T cells. (D) actTregs can colocalize with dendritic cells (DCs) and macrophages. Shown is 1 representative area from the image in panel A, with FOXP3 (green), CD11c (pink, DCs), CD68 (blue, macrophages), and CD21 (white). Arrow heads point at Tregs in proximity to DCs, whereas arrows point at Tregs in proximity to macrophages.
Treg subsets can be identified in external single-cell and bulk RNA-seq data sets, and have prognostic power in FL
Having shown that Treg subsets have distinct gene expression profiles (Figure 6A) and also differ in suppressive activity and abundance in tumor tissue, we then assessed their clinical relevance. To interrogate and enumerate the presence of Treg subsets across multiple gene expression data sets, both single-cell and bulk, we leveraged CIBERSORTx.37,38 We developed Treg-signature matrices using our scRNA-seq data as reference (supplemental Figure 10A-B; supplemental Table 9) and validated it for assigning Treg subset labels in scRNA-seq data (supplemental Figure 10C-D). We applied the resulting Treg-signature matrices to estimate the cellular abundances of resting, activated, and LAG3+ Tregs in 3 previously published scRNA-seq cohorts from reactive LN (n = 2), tonsils (n = 8), FL (n = 6), and DLBCL (n = 6).36,48,49 Remarkably, the distribution of the 3 Treg subsets in these external scRNA-seq data sets was highly concordant with the distribution in our scRNA-seq data (Pearson correlation, r = 0.96; Figure 6B-C; supplemental Figure 10E). Indeed, we confirmed that activated Tregs are more enriched in B-NHL than in reactive LN and tonsils. LAG3+ Tregs seemed to be associated with a non-neoplastic phenotype, because they were mainly found in tonsils and reactive LNs (Figure 6B; supplemental Figure 10E).
Validation of Treg clusters in separate scRNA-seq cohorts and prognostic significance by imputation of Treg subset frequencies in bulk RNA-seq cohorts. (A) Heat map of top 30 DEGs across the 3 Tregs subsets; actTregs, restTregs, and LAG3+ Tregs were plotted after normalization using scTransform. (B) Unique gene signature matrices were developed for CD4+ T-cell clusters including actTregs, restTregs, and LAG3+ Tregs, using the computational framework of CIBERSORTx. The matrices were then used to reannotate cell clusters in our scRNA-seq data sets and are shown as histograms of the frequency of Treg subsets in healthy donor tonsils (n = 3), FL (n = 3), and DLBCL (n = 3) (left); and in external scRNA-seq data sets (right), healthy donor tonsils (n = 8), FL (n = 7), and DLBCL (n = 6). (C) Pearson correlation of Treg subsets distribution in the external scRNA-seq data sets with the distribution in the scRNA-seq data generated in this study. (D) Abundance of the Treg subsets (left) and Kaplan-Meier survival curve for progression-free survival of patients with FL in the bulk RNA-seq cohort, stratified above the 85th percentile for the actTreg population (right).39 (E) Abundance of the Treg subsets (left) and Kaplan-Meier survival curve for failure-free survival of patients with FL in the external bulk RNA-seq cohort (right), in which patients were stratified using the same actTreg abundance threshold as in the discovery cohort (shown in panel D).40 ∗∗∗P < .001.
Validation of Treg clusters in separate scRNA-seq cohorts and prognostic significance by imputation of Treg subset frequencies in bulk RNA-seq cohorts. (A) Heat map of top 30 DEGs across the 3 Tregs subsets; actTregs, restTregs, and LAG3+ Tregs were plotted after normalization using scTransform. (B) Unique gene signature matrices were developed for CD4+ T-cell clusters including actTregs, restTregs, and LAG3+ Tregs, using the computational framework of CIBERSORTx. The matrices were then used to reannotate cell clusters in our scRNA-seq data sets and are shown as histograms of the frequency of Treg subsets in healthy donor tonsils (n = 3), FL (n = 3), and DLBCL (n = 3) (left); and in external scRNA-seq data sets (right), healthy donor tonsils (n = 8), FL (n = 7), and DLBCL (n = 6). (C) Pearson correlation of Treg subsets distribution in the external scRNA-seq data sets with the distribution in the scRNA-seq data generated in this study. (D) Abundance of the Treg subsets (left) and Kaplan-Meier survival curve for progression-free survival of patients with FL in the bulk RNA-seq cohort, stratified above the 85th percentile for the actTreg population (right).39 (E) Abundance of the Treg subsets (left) and Kaplan-Meier survival curve for failure-free survival of patients with FL in the external bulk RNA-seq cohort (right), in which patients were stratified using the same actTreg abundance threshold as in the discovery cohort (shown in panel D).40 ∗∗∗P < .001.
We, and others, have previously shown that the relative frequencies of cell subsets in the lymphoma microenvironment are prognostic.36,50 To investigate the clinical relevance of the Treg subsets identified here, we applied the Treg-signature matrices to FL cohorts with available bulk gene expression data. In our discovery cohort (supplemental Table 10),39 we noted a significantly increased frequency of activated Tregs among CD4+ T cells (Figure 6D), which was concordant with our cytometry data (Figure 4). Patients with FL with a high abundance of activated Tregs had significantly shorter progression-free survival in this cohort of 82 patients from 2 prospective trials with first-line rituximab treatment (P = .031; Figure 6D).39 Furthermore, high abundance of activated Tregs was significantly associated with inferior progression-free survival in unadjusted Cox regression analysis (hazard ratio [HR], 2.22; P = .035) and when adjusted for FL International Prognostic Index (FLIPI) (HR, 2.50; P = .018) and M7-FLIPI (HR, 3.11; P=.010) (supplemental Tables 11 and 12). In an independent FL cohort of 138 patients with FL treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (supplemental Table 13),40 high frequency of activated Tregs was also significantly associated with inferior failure-free survival (P = .013; Figure 6E). Furthermore, high frequency of activated Tregs was also significantly associated with failure-free survival in unadjusted Cox regression analysis (HR, 2.04; P = .015) and borderline significant when adjusted for M7-FLIPI, sex, age, and ECOG performance status (HR, 1.98; P = .055; supplemental Table 14). Of note, 23% of the patients in the validation cohort had missing information on M7-FLIPI and were excluded from the multivariable analysis. This loss of statistical power might have affected the borderline significant result in the adjusted analysis. These findings indicate that highly immunosuppressive Tregs have a negative impact on indolent lymphoma treated with rituximab with or without chemotherapy.
Discussion
In FL and DLBCL, activated Tregs was the major intratumoral Treg subset, characterized by high immunosuppressive capacity and coexpression of checkpoint receptors, distinguishing them from resting Tregs in the peripheral blood. Tregs in the TME with high suppressive activity represent a key challenge by limiting antitumor immunity. Higher frequency of Tregs is shown to render self-reactive CD8+ T cells hyporesponsive to antigen stimulation. These Treg-silenced CD8+ T cells express coinhibitory molecules such as CTLA4, produce very little cytokines, and have limited proliferation.51 Patients with B-NHL with relapsed/refractory disease are increasingly treated with immunotherapy, such as CAR T-cell therapy and bispecific antibodies.52-54 Support for lower efficacy of immunotherapy for patients with higher frequencies of tumor-infiltrating Tregs comes from a clinical trial in B-cell acute lymphoblastic leukemia, in which patients responding to the bispecific T-cell engager blinatumomab had lower frequencies of Tregs as compared with nonresponders.55 In patients with DLBCL treated with anti-CD19 CAR T cells (axicabtagene ciloleucel), high density of Tregs in pretreatment samples was not associated with response but severity of neurotoxicity.56 In contrast, high level of postinfusion CD19 CAR Tregs was associated with progressive disease.57 Treg diversity was not investigated in these studies but might further refine clinical associations.
Based on our scRNA-seq data, we leveraged CIBERSORTx to create CD4 signature matrices, and interrogated and validated the 3 Treg subsets in external lymphoma scRNA-seq cohorts, demonstrating remarkable similarity of the Treg subset abundances to our cohort. We identified the 3 subsets of Tregs in the scRNA-seq data set from Roider et al,48 who only reported 1 Treg cluster. Similarly, recent larger scRNA-seq studies in FL and DLBCL identified 1 Treg cluster.58-60 These studies sequenced all live cells from tumor samples in which tumor cells usually outnumbered other immune cells, whereas our strategy to sequence CD4 T cells facilitated deeper characterization of Tregs. Similar to our work, Aoki et al discovered 3 distinct transcriptional states of Tregs in Hodgkin lymphoma, all highly enriched as compared with reactive nonmalignant LNs.61 The authors described LAG3+ Tregs as the most abundant Treg subset in Hodgkin lymphoma tumors. In our study, LAG3+ Tregs accounted for a very small proportion of the tumor-infiltrating Tregs in FL and DLBCL but were abundant in nonmalignant tonsils and reactive LNs. We used our unique Treg signatures to impute the frequency of the different Treg subsets in 2 independent cohorts of FL with bulk gene expression data39,40 and found that a higher frequency of activated Tregs was associated with shorter progression-free survival in patients receiving rituximab or rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. This illustrates the clinical relevance of addressing Treg diversity in B-NHL.
In FL, activated Tregs were found in closer proximity to CD4+ and CD8+ T cells than other immune cells and tumor cells and were evenly distributed between the follicles and T-cell zones. Activated Tregs localizing to malignant follicles are likely follicular regulatory T (Tfr) cells, a specialized subset of Tregs first discovered in mice as CXCR5+BCL6+FOXP3+ T cells that localized to B-cell follicles and regulated Tfh-mediated B-cell responses after antigenic exposure.62,63 Tfr cells originate from thymic Tregs, with a TCR repertoire resembling that of Tregs.63,64 Evidence for Tfh origin has also been demonstrated, and the Tfh-to-Tfr conversion was important for germinal center regression.65 Our trajectory analysis suggested that activated Tregs were closest to resting Tregs, and these Treg subsets shared TCR clonotypes in FL. Tfr cells have earlier been studied in FL based on phenotypic identification as CD25+CXCR5hiICOShi cells, a Treg subset that is expanded in malignant LNs.66 CXCR5+ Tfr cells are found in tertiary lymphoid structures in several types of solid tumors and share clonotypes with CXCR5− intratumoral Tregs but demonstrate higher expansion.67 The authors further demonstrated that PD-1 checkpoint blockade is more potent if administered after Tfr cells had been depleted by anti-CTLA4, hence demonstrating that Tfr cells can negatively affect antitumor immunity.67
In-depth characterization of tumor-infiltrating Tregs with high suppressive capacity has the potential to uncover new strategies for selective targeting without causing systemic depletion of Tregs. Our study revealed that genes with significantly higher expression in activated Tregs as compared with resting Tregs include checkpoint receptors (TNFRSF4 and TNFRSF18), phosphatases (DUSP2 and DUSP4), NF-κB pathway molecules (NFKBIA, TNFAIP3, and NFKBIZ), chemokine receptors (CXCR4), and transcription factors (JUN, IRF1, and BATF). Global characterization of T cells in tumor tissue vs the peripheral blood and adjacent normal tissue, to our knowledge, has not yet been performed in B-NHL, but studies in non–small cell lung cancer and liver cancer identified a cluster of activated Tregs mainly found in tumor tissue.68,69 Meta-analysis across additional cohorts from melanoma, breast, colon, and lung cancers revealed a significant overlap of genes specifically expressed in tumor-infiltrating Tregs and identified a common signature of 31 genes including checkpoint receptors (CTLA4, TIGIT, TNFRSF4, TNFRSF18, and ICOS) and transcription factors (FOXP3, IKZF2, and BATF),68 corresponding well with the signature of activated Tregs in B-NHL. Targeting of Tregs is under active investigation, with most studies exploring various cell surface antigens as targets, including CD25, CD36, checkpoint receptors, and chemokine receptors.70 Targeting intracellular molecules in tumor-infiltrating Tregs might also be a powerful strategy. Here, deletion or inhibition with chemical compounds targeting NF-κB pathway molecules such as c-Rel and CARD11/CARMA1 suppressed Treg activity and potentiated the effect of checkpoint blockade in vivo.71,72 This is interesting in relation to our observation that NF-κB pathway genes are upregulated in activated Tregs in B-NHL tumors.
To generate effective antitumor immunity, a key goal is to target markers preferentially expressed by intratumoral Tregs and not by systemic Tregs. Our in-depth characterization of Tregs in B-NHL tumors and healthy tissue have illuminated transcriptional and proteomic differences between Treg subsets and divergent frequencies dependent on tissue localization. This may translate into selective targeting of activated Tregs, the dominant suppressor of tumor immunity, thereby enable safer and more potent cancer immunotherapy.
Acknowledgments
The authors acknowledge Vera Hilden for the technical support and the support staff at the Flow Cytometry Core Facility and the Genomics Core Facility at Institute for Cancer Research, Oslo University Hospital.
This work was funded by the KG Jebsen Foundation, Centre for B-cell Malignancies (centre no. 19) (E.B.S. and J.H.M.), the Norwegian Cancer Society (162948 [K.H.], 182694 [E.B.S.], and 245270 [J.H.M.]), the Research Council of Norway (FRIMEDBIO 230817/F20) (E.B.S.), and the American Association for Cancer Research (19-40-12-STEE) (C.B.S.), and was partially supported by the Research Council of Norway through its Centres of Excellence scheme, project number 332727.
Authorship
Contribution: I.S., S.E.J., E.B.S., K.H., and J.H.M. conceptualized the study; I.S., A.S., C.B.S., S.E.J., Y.N.B., K.I., K.H., and J.H.M. were responsible for formal analysis; I.S., A.S., C.B.S., and K.H. were responsible for validation; I.S., A.S., C.B.S., Y.N.B., M.S.F., S.L., and K.H. performed the investigation; I.S., A.S., C.B.S., S.E.J., Y.N.B., A.K., K.H., H.H., B.Ø., M.B., and E.K. were responsible for data curation; I.S., A.S., C.B.S., S.E.J., A.A.A., K.H., and J.H.M. were responsible for visualization; I.S., A.S., C.B.S., S.E.J., Y.N.B., E.H.R., S.M., S.C., K.K., M.S.F., K.T., A.M.N., A.A.A., K.H., and J.H.M. were responsible for the methodology; I.S. and J.H.M. wrote the original draft of the manuscript; I.S., A.S., C.B.S., S.E.J., Y.N.B., J.O., K.T., S.L., E.B.S., A.A.A., K.H., and J.H.M. reviewed and edited the manuscript; I.S., A.S., C.B.S., E.H.R., A.M.N., and A.A.A. were responsible for software; A.K., K.B., S.S., H.H., B.Ø., M.B., E.K., J.O., and J.H.M. provided resources; E.B.S., K.H., A.A.A., and J.H.M. were responsible for funding acquisition; K.H. and J.H.M. supervised the study; J.H.M. was responsible for project administration; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: H.H. is a member on the advisory committees for Gilead, Roche, Nanovector, Novartis, Nordic, and Takeda. A.A.A. is a member of the Cell advisory board; reports research support from Bristol Myers Squibb; reports ownership interest in CiberMed, FortySeven Inc, and Foresight Diagnostics; reports patent filings related to cancer biomarkers; and reports paid consultancy for Genentech, Roche, Chugai, Gilead, and Celgene. A.M.N. reports ownership interest in CiberMed and patent filings related to cancer biomarkers. C.B.S., A.A.A., and A.M.N. have filed a patent application (PCT/US2020/059196). The remaining authors declare no competing financial interests.
Correspondence: June H. Myklebust, Department of Cancer Immunology, Institute for Cancer Research, Oslo University Hospital, Postboks 4953 Nydalen, 0424 Oslo, Norway; e-mail: j.h.myklebust@medisin.uio.no.
References
Author notes
∗A.S. and C.B.S. contributed equally to this study.
Presented in abstract form at the 62nd annual meeting of the American Society of Hematology (virtual), 5-8 December 2020, and the 63rd annual meeting of the American Society of Hematology, Atlanta, GA, 11 December 2021.
Single-cell RNA-seq data are deposited in the Gene Expression Omnibus (accession number GSE242756).
We used ShinyCell1 to generate an interactive portal for exploration of the single-cell RNA-seq data, available at https://myklebust.medisin.uio.no/scrna/treglymphoma/. The scripts are available at github.com/myklebustlab/TregLymphoma.
The full-text version of this article contains a data supplement.