Key Points
Age at the time of exposure influences the outcome of PRR engagement, and this biology is shared between mouse and human B-ALL.
TLR3 engagement by pathogenic dsRNA induces tumor necrosis factor α–dependent preleukemia cell proliferation in early life.
Abstract
Common infections have long been proposed to play a role in the development of pediatric B-cell acute lymphoblastic leukemia (B-ALL). However, epidemiologic studies report contradictory effects of infection exposure on subsequent B-ALL risk, and no specific pathogen has been definitively linked to the disease. A unifying mechanism to explain the divergent outcomes could inform disease prevention strategies. We previously reported that the pattern recognition receptor (PRR) ligand Poly(I:C) exerted effects on B-ALL cells that were distinct from those observed with other nucleic acid–based PRR ligands. Here, using multiple double-stranded RNA (dsRNA) moieties, we show that the overall outcome of exposure to Poly(I:C) reflects the balance of opposing responses induced by its ligation to endosomal and cytoplasmic receptors. This PRR response biology is shared between mouse and human B-ALL and can increase leukemia-initiating cell burden in vivo during the preleukemia phase of B-ALL, primarily through tumor necrosis factor α signaling. The age of the responding immune system further influences the impact of dsRNA exposure on B-ALL cells in both mouse and human settings. Overall, our study demonstrates that potentially proleukemic and antileukemic effects can each be generated by the stimulation of pathogen recognition pathways and indicates a mechanistic explanation for the contrasting epidemiologic associations reported for infection exposure and B-ALL.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL), the most common childhood malignancy,1,2 arises from the clonal expansion of abnormal precursor B (BCP) cells. In most B-ALL cases, BCP cells acquire cytogenetic abnormalities (such as aneuploidy or chromosomal translocations) in utero,3-5 which confer developmental arrest and survival and/or proliferative advantages.6 Although necessary for leukemogenesis, these early occurring genomic abnormalities are not sufficient to drive full BCP cell transformation and instead give rise to a clinically silent preleukemic phase characterized by a low burden of potential leukemia-initiating cells (LICs).5 Notably, detection of B-ALL–initiating genomic lesions in infants is significantly more common than the incidence of B-ALL,3,7,8 and the postnatal acquisition of additional oncogenic events is essential to the emergence of overt leukemia.9,10
Second hit mutations associated with the transition from preleukemia to leukemia have been identified,10-12 but the determinants of disease progression remain poorly defined. A role for infections in the etiology of pediatric B-ALL has long been hypothesized, and epidemiological studies have provided evidence that infection exposure, especially early in life, affects B-ALL risk. Although several studies indicated a protective influence when surrogates of infection exposure, such as daycare attendance, breastfeeding, and mode of delivery, were analyzed,13-16 other studies have reported that infections are associated with an increased risk of B-ALL.17-19 Similarly, intriguing, but not definitive, data exist for the protective effects of early vaccination.20,21 These findings have provided support for the prominent delayed infection hypothesis, which posits that childhood B-ALL is, in part, fueled by a lack of early life immune stimulation that leads to a dysregulated, leukemia-promoting immune response to later common infections.22,23
The nature of the early life immune modulation and identity of microbial agents that influence B-ALL progression remain unknown. In the absence of clear associations between specific pathogens and B-ALL risk, timing of infection exposure has emerged as a key outcome variable.22 In addition, an influence of early life microbial colonization on B-ALL progression has been suggested from mouse model studies.24 Because viruses are both significant components of the early microbiome25 and pathogens associated with childhood B-ALL progression,26-28 here we examined the response of B-ALL cells to double-stranded RNA (dsRNA), which is generated during infections by most families of virus.29 Our results reveal that virus-associated signaling can induce either expansion or depletion of human B-ALL as well as murine B-ALL and LICs, with the outcome being influenced by the specific pattern recognition receptor (PRR) pathways activated and the maturity of the responding immune system.
Methods
Mice
The Eμ-Ret mouse model of precursor B-ALL (generously provided by Stephan Grupp, University of Pennsylvania, Philadelphia, PA) was maintained on a BALB/c background (Jackson Laboratory, Bar Harbor, ME) through in-house breeding.30 Both male and female mice were used in all experiments. Primary human B-ALL cells were freshly isolated from patient-derived xenografts (PDXs) established by tail vein injection of diagnostic B-ALL bone marrow (BM) samples in NOD.Cg-Prkdcscid/IL2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory). Fully engrafted mice were euthanized, and human B-ALL cells were isolated from the spleen and BM. Experiments were conducted in accordance with a University of British Columbia Animal Care Committee–approved protocol (A19-0197).
Patient-derived B-ALL samples
Cryopreserved BM samples, collected at the time of initial diagnosis from pediatric patients with B-ALL, were obtained from the BC Children’s Hospital Biobank. Clinical characteristics for each patient are shown in supplemental Table 1. Informed consent from human participants was obtained in all cases in accordance with the Declaration of Helsinki, and the study performed under a University of British Columbia Children’s & Women’s Research Ethics Board-approved protocol (H17-01860).
Flow cytometry
Single-cell suspensions from neonate and adult mouse spleen and BM were stained with the following fluorochrome-conjugated antibodies: B220 (RA3-6B2), BP-1(6C3), CD43(S7), CD24 (M1/69), CD19 (6D5), CD117 (ACK2), CD25 (PC61), immunoglobin (IgM [RMM-1]; BioLegend, San Diego, CA), and CD43(S7) (BD Biosciences, San Jose, CA). For in vitro assays, Eμ-Ret LIC and B-ALL cells were stained using B220 (RA3-6B2), BP-1 (6C3), CD19 (1D3), and 7-AAD (BioLegend). E2A-PBX1 transgenic mouse-derived leukemia cells were identified based on their green fluorescent protein expression.31 B-ALL cells from patients were identified using fluorochrome-conjugated antibodies for CD19 (HIBI9), CD45 (HI30), HLA-A,B,C (W6/32), and 7-AAD (BioLegend). Data from in vitro assays were acquired on Accuri C6 or FACSymphony A1 cytometers (BD Biosciences), whereas data from in vivo experiments were acquired on a BD LSRII or BD Fortessa cytometer (BD Biosciences). Data were analyzed using FlowJo software v10.8 (BD Bioscience) using established gating strategies (supplemental Figures 1 and 2). For all experiments, specific cell subsets were quantified using CountBright Absolute Counting Beads (Invitrogen, Carlsbad, CA). For flow cytometric sorting of LICs, single-cell suspensions from Eμ-Ret mouse spleen or BM were stained with fluorochrome-conjugated antibodies for B220 (RA3-6B2), BP-1 (6C3), CD19 (1D3), and 7-AAD (BioLegend). LICs (CD19+/B220int/BP-1hi) were purified using MoFlo Astrios EQ cell sorter (Beckman Coulter Inc, Brea, CA).
In vitro experiments
A variety of B-ALL cell types were used in this study: (1) Eμ-Ret LIC (flow-purified based on their B220int/BP-1hi/CD19+ phenotype or maintained in bulk cultures) were isolated from the spleen and BM of healthy Eμ-Ret mice; (2) primary mouse-derived leukemic B-ALL cells were harvested from spleens of moribund Eμ-Ret and Tg(E2A-PBX1)Pax+/−Cd19-Cre mice (abbreviated as E2A-PBX1), as described previously24,31; and (3) primary human B-ALL cells were harvested from spleens of PDXs when they reached a high burden of leukemia.
LICs or B-ALL cells were cultured at 2.5 × 106/mL in the presence or absence of 10 μg/mL of dsRNA mimics, polyinosinic-polycytidylic acid (Poly(I:C)), polyadenylic-polyuridylic acid (Poly(A:U)), and 5′ triphosphate dsRNA (5′ppp-dsRNA) (Invivogen, San Diego, CA). The number of viable cells was assessed after 48 hours by flow cytometry. Apoptosis was measured using annexin V binding kits (BioLegend), following the manufacturer’s protocol. For assessment of proliferation, 10 μM 5-bromo-2′-deoxyuridine (BrdU) was added at the initiation of the culture and incorporation measured by flow cytometry using a BrdU detection kit (BD Biosciences), following the manufacturer’s instructions. Graphs depict pooled results from at least 2 independent experiments.
To identify immune cell subsets and sources of tumor necrosis factor α (TNF-α), splenocytes from 6-day-old (pup) and 4- to 6-week-old (adult) mice were stimulated with Poly(A: U) or left unstimulated for 16 hours at 37°C. After stimulation, cells were treated with 3 μg/mL Brefeldin A solution (BioLegend) for 4 hours. Cells were harvested and surface stained with the following fluorochrome-conjugated antibodies: B220 (RA3-6B2), CD19 (1D3), CD3 (17A2), F4/80 (BM8), CD49b (DX5) (BioLegend), CD4 (GKI.5), CD8 (53-6.7), CD11b (M1/70), CD11c (B-ly6), CD335 (29A1.4), and major histocompatibility complex class 2 (MHC CII [2G9]; BD Biosciences). Zombie Aqua fixable viability dye (BioLegend) was used to exclude dead cells. Cells were fixed and permeabilized using eBioscience Intracellular Fixation & Permeabilization Buffer Set (Invitrogen) following the manufacturer’s instructions. Cells were then stained for anti-mouse TNF-α (MP6T22) (BioLegend), and data were analyzed using established gating strategies (supplemental Figures 3 and 4).
For mouse coculture experiments, 6 × 104 purified Eμ-Ret LICs were incubated with 2 × 106 bulk splenocytes or BM cells from 1- to 2-week-old or 4- to 6-week-old wild-type BALB/c mice. For human coculture experiments, purified PDX B-ALL cells were labeled with CellTrace carboxyfluorescein diacetate succinimidyl ester (Thermo Fisher Scientific, Waltham, MA) and then incubated with cord blood mononuclear cells or adult peripheral blood mononuclear cells (PBMCs), obtained from the BC Children’s Hospital Biobank, at an effector-to-target ratio of 20:1. Both mouse and human cocultures were treated with or without dsRNA, and the number of viable cells were quantified after 48 hours using CountBright Absolute Counting Beads. For human B-ALL cells, carboxyfluorescein diacetate succinimidyl ester–positive cells were measured.
In vivo experiments
Eμ-Ret mice were treated with the indicated PRR ligands by intraperitoneal injection. Neonates received 10 μg per injection of indicated PRR ligands, whereas adult mice received 50 μg. Mice were euthanized 7 days later, and LIC populations in the spleen and BM were assessed by flow cytometry, based on Hardy fraction classification (supplemental Figure 1).32 For detection of proliferation, Eμ-Ret mice injected 4 days previously with Poly(A:U) or phosphate-buffered saline were injected with 0.5 mg of BrdU (BD Biosciences). After 2 days, spleen and BM were harvested, and BrdU incorporation by LICs was measured by flow cytometry using a BrdU detection kit (BD Biosciences). For cytokine neutralization experiments, neonate mice received 50 μg of anti–TNF-α (clone XT3.11; rat IgG1) or anti–interleukin-10 (IL-10) (clone JES-2A5; rat IgG1) antibody (Bio X Cell, Lebanon, NH), followed by a 10 μg dose of Poly(A:U) after 30 minutes. Mice were euthanized 7 days later, and LIC populations in the spleen and BM were quantified. The number of LIC in each spleen was quantified for all experiments using CountBright Absolute Counting Beads (Invitrogen).
RNA sequencing
Eμ-Ret pups were treated with either phosphate-buffered saline or Poly(A:U), and LICs were purified 7 days later using a MoFlo Astrios EQ cell sorter (Beckman Coulter Inc). RNA was isolated from sorted LICs using AllPrep DNA/RNA Mini kit (Qiagen, Hilden, Germany). Library preparation and RNA sequencing was performed at the Biomedical Research Centre at the University of British Columbia, Vancouver. Raw count matrix was processed with DESeq233 Bioconductor package within the R Studio programming environment. Low-count (total count = 0) genes were removed from data sets and then normalized using DESeq2 median of ratios method. Genes with sum of averages of normalized data ≤20 in experimental groups were filtered out. For gene set enrichment analysis (GSEA),34 normalized counts were compared with the hallmark gene sets of the Molecular Signature Database.35
Cytokine and chemokine analysis
The serum was collected at the experimental end point, 7 days after PRR ligand treatment. Cytokines and chemokines were detected in the serum samples using LegendPLEX mouse cytokine and proinflammatory chemokine 13-plex assay kits (BioLegend), according to the manufacturer’s instructions.
Statistical methods
GraphPad Prism 9 for Mac OS software (GraphPad, San Diego, CA) was used for all statistical analysis. Analyses of all in vivo and in vitro cell survival assays were performed using 1-way analysis of variance (Dunnett multiple comparisons test). Two-way analysis of variance was used when >1 variable was involved (eg, in vitro age-dependency assay). All graphs represent at least 2 independent repeats. Specific P values for each experiment are shown in figure legends.
Results
dsRNA-specific PRRs induce differential responses in both murine and patient B-ALL cells
We have previously shown that ligands for the toll-like receptor (TLR) family of PRR can influence LIC survival and B-ALL progression in Eμ-Ret mice.36,37 In contrast to other nucleic acid–based TLR ligands, which consistently induce depletion of LIC and B-ALL cells, Poly(I:C) produced variable responses, including modest population expansion in some settings. Poly(I:C) is a synthetic dsRNA that binds to TLR3 in endosomes as well as to retinoic-acid-inducible gene I (RIG-I) and the melanoma differentiation–associated gene 5 receptors in the cytosol.38,39 TLR3 and RIG-I are both key sensors of viral infection, leading to type 1 interferon (IFN) production and host immune responses to viruses. To further investigate the variable B-ALL outcomes induced by Poly(I:C), we first assessed the effects of ligands that are specific for distinct receptors that bind Poly(I:C); namely, 5′ppp-dsRNA, a RIG-I agonist, and Poly(A:U), a TLR3 agonist.40 Consistent with our previous studies, we observed a trend toward modest increases in viable Eμ-Ret leukemia cell numbers after incubation with Poly(I:C) (fold change, 1.2; P > .05; Figure 1A). In contrast to this marginal effect, leukemia cell numbers were significantly increased by Poly(A:U) (fold change, 1.7; P < .0001) and significantly reduced by 5′ppp-dsRNA (fold change, 0.5; P < .001), indicating divergent responses to the 2 dsRNA moieties. To identify the cell responses underlying the increase in Eμ-Ret B-ALL numbers after treatment with Poly(A:U), we assessed cell death and proliferation (Figure 1B; supplemental Figure 5). An increase in BrdU+ B-ALL cells was concurrent with a reduction in the number of dead cells. No change in apoptosis levels was detected. These results suggest that the increase in blast cell number reflects augmented survival and proliferation.
Distinct dsRNA moieties induce differential responses in both murine and human B-ALL cells and LICs in vitro. (A) B-ALL cells from Eμ-Ret mice were stimulated with the indicated ligands or left unstimulated and viable cells number measured after 48 hours. Results are pooled from at least 5 individual experiments. (B) Cultures of leukemia cells isolated from Eμ-Ret mice were assessed for live (annexin-negative and 7AAD-negative), dead (annexin-positive and 7AAD-positive), apoptotic (annexin-positive and 7AAD-negative), and proliferating (BrdU+) cell characteristics after 48 hours in the presence or absence of Poly(A:U). Results are pooled from 3 individual experiments. (C) E2A-PBX1, (D) patient-derived B-ALL leukemic blasts, and (E) purified LICs from the spleen and BM of 2- to 8-week-old Eμ-Ret mice were exposed to Poly(A:U), Poly(I:C), or 5′ppp-dsRNA for 48 hours, and the number of viable cells was measured. Results are pooled from at least 5 individual experiments. Individual patient samples are color-coded in panel D. In all experiments, cell counts were quantified using CountBright beads. Dunnett multiple comparison test; bars represent mean ± standard deviation (SD); ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant.
Distinct dsRNA moieties induce differential responses in both murine and human B-ALL cells and LICs in vitro. (A) B-ALL cells from Eμ-Ret mice were stimulated with the indicated ligands or left unstimulated and viable cells number measured after 48 hours. Results are pooled from at least 5 individual experiments. (B) Cultures of leukemia cells isolated from Eμ-Ret mice were assessed for live (annexin-negative and 7AAD-negative), dead (annexin-positive and 7AAD-positive), apoptotic (annexin-positive and 7AAD-negative), and proliferating (BrdU+) cell characteristics after 48 hours in the presence or absence of Poly(A:U). Results are pooled from 3 individual experiments. (C) E2A-PBX1, (D) patient-derived B-ALL leukemic blasts, and (E) purified LICs from the spleen and BM of 2- to 8-week-old Eμ-Ret mice were exposed to Poly(A:U), Poly(I:C), or 5′ppp-dsRNA for 48 hours, and the number of viable cells was measured. Results are pooled from at least 5 individual experiments. Individual patient samples are color-coded in panel D. In all experiments, cell counts were quantified using CountBright beads. Dunnett multiple comparison test; bars represent mean ± standard deviation (SD); ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant.
Although Eμ-Ret mice exclusively develop B-ALL, Ret is not a recognized driver of B-ALL in children. To address the possibility that the observed effect of dsRNA analogs is specific to Ret fusion gene-driven leukemia, we exposed B-ALL cells derived from E2A-PBX1 mice to the dsRNA mimics.31 The same response pattern as in Eμ-Ret B-ALL was observed with E2A-PBX1 leukemia, with Poly(A:U) increasing B-ALL cell numbers (fold change, 1.3; P < .001), whereas 5′ppp-dsRNA reduced them (fold change, 0.5; P < .0001; Figure 1C). Furthermore, because mouse B-ALL may not share identical biology with the human disease, we treated B-ALL cells from 6 different patients similarly (Figure 1D). An identical response pattern was observed, with Poly(I:C) responses appearing to reflect the opposing effects of Poly(A:U) (fold change, 1.6; P < .0001) and RIG-I (fold change, 0.5; P < .0001). These results indicate that the divergent direct effects of dsRNA-specific PRR engagement are shared across different B-ALL subtypes and between human and mouse leukemia.
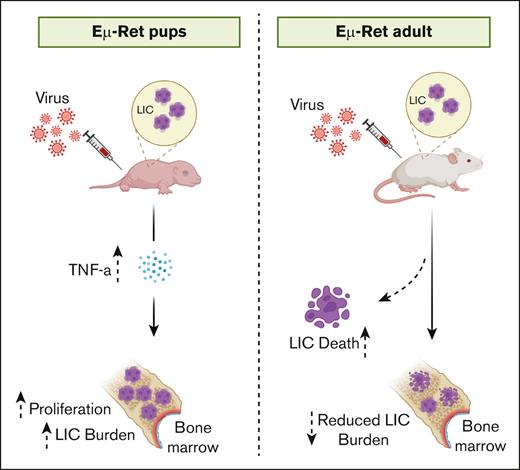
Evidence suggests that infection exposure during the preleukemic phase affects B-ALL progression. Given the many challenges of studying the preleukemic phase in infants, we evaluated the impact of pathogenic dsRNA on LIC isolated from healthy Eμ-Ret mice. In Eμ-Ret mice, B-ALL develops from an in utero–generated preleukemic BCP population after a variable latency of 3 to 12 months.30,41 Purified LIC from the spleen and BM of healthy mice were exposed to dsRNA mimics for 48 hours, and the number of viable cells was measured (Figure 1E). As was seen for fully transformed leukemia cells, 5′ppp-dsRNA reduced viable LIC recovery (spleen fold change, 0.7; not significant; and BM fold change, 0.5; P < .01), whereas Poly(A:U) treatment resulted in significantly increased LIC numbers (spleen fold change, 1.5; P < .001; and BM fold change, 1.7; P < .0001). Poly(I:C) induced only marginal expansion (spleen fold change, 1.2; not significant; and BM fold change, 1.3; P < .05). The Poly(A:U)-induced increase in cell number was accompanied by the same trend of increased proliferation and reduced death observed with Eμ-Ret leukemia cells (supplemental Figure 6). This result indicates that the PRR responses observed for B-ALL cells are not a result of full leukemic transformation, and such signals could exert an impact during the preclinical phase of the disease.
Age of immune effectors influences the outcome of TLR3 engagement
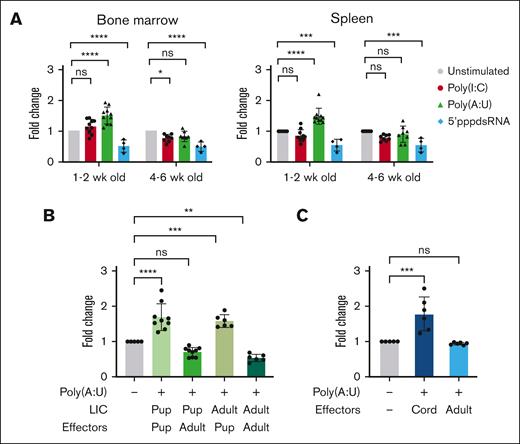
In addition to potential direct effects on B-ALL cells, exposure to TLR ligands will induce immune activity that can result in the depletion of B-ALL cells.37,42 Because newborns have developing immune systems,43-45 age-related changes in response to infectious agents could affect their impact on preleukemic BCP cells. In addition to the obvious differences in the total cell numbers, neonate and adult spleens show significant differences in the proportions of primary immune subsets (supplemental Figure 7). To assess the influence of immune system maturity on distinct LIC outcomes after dsRNA exposure, we treated bulk spleen and BM cells from 1- to 2- and 4- to 6-week-old Eμ-Ret mice with dsRNA mimics (Figure 2A). In neonatal bulk organ cultures, Poly(A:U), but not Poly(I:C), significantly increased LIC numbers (spleen fold change, 1.5; P < .001; and BM fold change, 1.5; P < .001). However, this expansion effect was not observed in cultures established from older mice (spleen fold change, 0.9; ns; and BM fold change, 0.8; ns). In contrast, bulk organ cultures established from both neonatal and adult mice significantly depleted LIC in response to 5′ppp-dsRNA (spleen fold change, 0.5; P < .001; and BM fold change, 0.5, P < .001 for neonate cultures; and spleen fold change, 0.5; P < .001; and BM fold change, 0.5; P < .001 for adult cultures), with no difference in the magnitude of effect observed between settings. These results indicate that some, but not all, pathogen response pathways show age-related differences.
The age of immune effectors determines the outcome of TLR3 engagement. (A) Bulk splenocytes and BM cells from healthy Eμ-Ret mice of varying ages were exposed to indicated ligands and viable LIC recovery measured after 48 hours. Results are pooled from at least 4 individual experiments and analyzed using Dunnett multiple comparison test. (B) Purified LIC from neonatal and adult Eμ-Ret mice were incubated alone or with splenocytes from 2-wk-old (neonate) or 4-wk-old (adult) BALB/c mice. Cultures were treated with Poly(A:U), and viable cell recovery was measured after 48 hours and compared with that of unstimulated controls. Results are pooled from at least 3 individual experiments and analyzed using Dunnett multiple comparison test. (C) Four human B-ALL samples were incubated with cord or adult PBMCs. Cultures were treated with or without Poly(A:U), and viable cell recovery was measured using CountBright beads after 48 hours and analyzed using unpaired t test. For all graphs, bars represent mean ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns. wk, week.
The age of immune effectors determines the outcome of TLR3 engagement. (A) Bulk splenocytes and BM cells from healthy Eμ-Ret mice of varying ages were exposed to indicated ligands and viable LIC recovery measured after 48 hours. Results are pooled from at least 4 individual experiments and analyzed using Dunnett multiple comparison test. (B) Purified LIC from neonatal and adult Eμ-Ret mice were incubated alone or with splenocytes from 2-wk-old (neonate) or 4-wk-old (adult) BALB/c mice. Cultures were treated with Poly(A:U), and viable cell recovery was measured after 48 hours and compared with that of unstimulated controls. Results are pooled from at least 3 individual experiments and analyzed using Dunnett multiple comparison test. (C) Four human B-ALL samples were incubated with cord or adult PBMCs. Cultures were treated with or without Poly(A:U), and viable cell recovery was measured using CountBright beads after 48 hours and analyzed using unpaired t test. For all graphs, bars represent mean ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns. wk, week.
The previous experiment indicated that Poly(A:U) failed to induce LIC expansion in bulk organ cultures from older mice. Because the immune system and LIC population are both older in that setting, the precise nature of this age dependency is unclear; intrinsic age-related changes may render LIC more or less sensitive to immune signals. To clarify which cell population confers the age dependency, purified LIC from neonatal and adult Eμ-Ret mice were cultured in the presence of splenocytes from 2-week-old or 4-week-old BALB/c mice, with or without Poly(A:U), for 48 hours (Figure 2B). An expansion of viable LIC numbers was achieved by Poly(A:U) treatment in the presence of neonatal splenocytes, irrespective of the age of the LIC (neonate LIC fold change, 1.7; P < .001; and adult LIC fold change, 1.6; P < .001). In contrast, the same stimulation in the presence of 4-week-old splenocytes resulted in reduced numbers of both neonate- and adult-derived LIC (neonate fold change, 0.7; P < .05; and adult fold change, 0.5; P < .001).
To evaluate the potential impact of immune system maturation in the context of human B-ALL, we cocultured B-ALL cells from patients with cord blood mononuclear cells or adult PBMCs, in the presence or absence of Poly(A:U), for 48 hours (Figure 2C; supplemental Figure 8). Consistent with results for Eμ-Ret LIC, Poly(A:U) treatment in the presence of cord blood cells increased the number of viable B-ALL cells (fold change, 1.8; P < .001). No such expansion was observed in cocultures with adult PBMC (fold change, 0.9; ns). These results implicate maturation of the immune environment as the determinant of LIC fate after immune modulation, in both human and mouse settings.
Poly(A:U)-induced LIC proliferation in vivo is an age-dependent outcome
In vitro culture in the presence of PRR ligands exposes B-ALL cells to direct stimulation that may not occur in vivo. Therefore, we next investigated whether similar responses were obtained in vivo. We treated neonatal (aged 1 week) and adult (aged 4-6 weeks) Eμ-Ret mice with Poly(I:C), Poly(A:U), and/or 5′ppp-dsRNA and quantified LIC numbers 7 days after treatment. Although 5′ppp-dsRNA (spleen fold change, 0.4; ns; and BM fold change, 0.4; ns) and Poly(I:C) (spleen fold change, 0.9; ns; and BM fold change, 0.7; ns) led to varying levels of LIC depletion in neonatal mice, treatment with Poly(A:U) generated a significant increase in LIC numbers (spleen fold change, 1.9; P < .001; and BM fold change, 1.8; P < .01; Figure 3A). As would be hypothesized based on the consistently opposing effects of the TLR3 and RIG-I ligands on the LIC population, administration of a combined dose of Poly(A:U) and 5′ppp-dsRNA yielded a similar outcome to that seen with Poly(I:C), with little change in the size of the LIC population (spleen fold change, 1.1; ns; and BM fold change, 0.9; ns). In adult Eμ-Ret mice, however, Poly(A:U) did not induce LIC expansion but instead caused a significant reduction in LIC numbers (spleen fold change, 0.4; P < .001; and BM fold change, 0.6; P < .001) (Figure 3B). These results confirm the influence of age on LIC response in the setting of dsRNA-induced immune responses in vivo.
Poly(A:U)-induced LIC expansion in vivo is an age-dependent outcome. (A) 6-day-old Eμ-Ret pups were injected with indicated PRR ligands, and LIC burden was evaluated 7 days later and analyzed using Dunnett multiple comparison test. (B) 4- to 6-week-old Eμ-Ret mice were treated with either phosphate-buffered saline (PBS) or Poly(A:U), and LIC burden after 7 days was evaluated. (n = 5 for PBS and n = 5 for Poly(A:U)). In all experiments, cell counts were quantified using CountBright beads. Unpaired t test; bars represent mean ± SD; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns.
Poly(A:U)-induced LIC expansion in vivo is an age-dependent outcome. (A) 6-day-old Eμ-Ret pups were injected with indicated PRR ligands, and LIC burden was evaluated 7 days later and analyzed using Dunnett multiple comparison test. (B) 4- to 6-week-old Eμ-Ret mice were treated with either phosphate-buffered saline (PBS) or Poly(A:U), and LIC burden after 7 days was evaluated. (n = 5 for PBS and n = 5 for Poly(A:U)). In all experiments, cell counts were quantified using CountBright beads. Unpaired t test; bars represent mean ± SD; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns.
To assess whether the BCP cell expansion induced by Poly(A:U) treatment of neonates was limited to LIC or was the result of a broader stimulatory activity, we administered Poly(A:U) to 6-day-old Eμ-Ret pups and compared BCP cell populations in the BM and spleen with nontreated controls using flow gating to define Hardy fractions.32 Increases in cell number in response to Poly(A:U) treatment were restricted to fraction C (the late pro–B-cell fraction that contains LIC) and the mature B-cell population (Figure 4A and B; fraction C fold change, 1.6; P < .05; and B-cell fold change, 1.9; P < .001). Expansion of pre–pro-B and early pro-B populations (fractions A and B) was not detected. Consistent with this result, there was a higher percentage of BrdU+ LIC and B cells in the spleen and BM of Poly(A:U)-treated mice, implicating increased proliferation as a driver of elevated cell numbers (Figure 4C-D). This result indicates that Poly(A:U)-induced expansion is not restricted to LIC but also reflects a normal physiologic response within the B-cell compartment.
Poly(A:U) induces LIC expansion in vivo. (A) Representative flow plots depicting early precursor B-cell populations (Hardy fractions [Fr] A, B, and C/C′) from the spleen and BM of PBS- or Poly(A:U)-treated mice. (B) FrA, FrB, and FrC/C′ and mature B cells (B cell) were quantified by flow cytometry from PBS- and Poly(A:U)-treated mice. Cell counts were quantified using CountBright beads and results pooled from at least 3 individual experiments (Dunnett multiple comparison test). (C) Representative flow cytometry plots depicting BrdU+ LIC in the spleen and BM from PBS- or Poly(A:U)-treated mice. (D) Early BCP proliferating in the BM of Poly(A:U)-treated compared with PBS-treated control mice, expressed as % BrdU+ B220+CD43+ cells. Results are pooled from 2 individual experiments (Šídák multiple comparisons test). Bars represent mean ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns.
Poly(A:U) induces LIC expansion in vivo. (A) Representative flow plots depicting early precursor B-cell populations (Hardy fractions [Fr] A, B, and C/C′) from the spleen and BM of PBS- or Poly(A:U)-treated mice. (B) FrA, FrB, and FrC/C′ and mature B cells (B cell) were quantified by flow cytometry from PBS- and Poly(A:U)-treated mice. Cell counts were quantified using CountBright beads and results pooled from at least 3 individual experiments (Dunnett multiple comparison test). (C) Representative flow cytometry plots depicting BrdU+ LIC in the spleen and BM from PBS- or Poly(A:U)-treated mice. (D) Early BCP proliferating in the BM of Poly(A:U)-treated compared with PBS-treated control mice, expressed as % BrdU+ B220+CD43+ cells. Results are pooled from 2 individual experiments (Šídák multiple comparisons test). Bars represent mean ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns.
TNF-α contributes to LIC expansion in neonates
Our results reveal distinct LIC responses to changes in their immune environment during early life. To identify the specific components of the neonatal Poly(A:U) response that altered LIC physiology, we performed RNA sequencing on LIC purified from control and treated Eμ-Ret pups. GSEA revealed that 6 of 48 hallmark pathways were significantly upregulated, whereas 14 of 48 pathways were significantly downregulated in Poly(A:U)-treated LICs (Figure 5A; supplemental Figures 9 and 10). Notably, differentially expressed genes showed significant enrichment for TNF-α signaling (false discovery rate [FDR] q value = 0.003), with concurrent downregulation of IFN-α (FDR q value = 0.0) and IFN-γ (FDR q value = 0.0) response signaling (Figure 5B). Furthermore, the upregulation of Myc target genes is consistent with the observed increase in LIC proliferation after exposure to Poly(A:U). Myc is a critical driver of normal pro–B-cell expansion.46 Consistent with the TNF-α signaling detected in GSEA, TNF-α was the most significantly elevated cytokine in treated neonates (P < .01), but it was not detected in adults (Figure 5C; supplemental Figure 11).
RNA-seq reveals gene expression changes in LIC after exposure to Poly(A:U). (A) GSEA showing upregulated (top) and downregulated (bottom) pathways in LICs from Poly(A:U)-treated young Eμ -Ret mice compared with those in LICs from untreated littermates. (B) Enrichment plot of genes in significantly altered hallmark pathways gene sets. (C) Plasma cytokine levels measured 16 hours after PBS or Poly(A:U) administration to Eμ-Ret neonates and adults (n = 3 mice per group). MCP-1, (Monocyte Chemoattractant Protein-1).
RNA-seq reveals gene expression changes in LIC after exposure to Poly(A:U). (A) GSEA showing upregulated (top) and downregulated (bottom) pathways in LICs from Poly(A:U)-treated young Eμ -Ret mice compared with those in LICs from untreated littermates. (B) Enrichment plot of genes in significantly altered hallmark pathways gene sets. (C) Plasma cytokine levels measured 16 hours after PBS or Poly(A:U) administration to Eμ-Ret neonates and adults (n = 3 mice per group). MCP-1, (Monocyte Chemoattractant Protein-1).
To identify the source of TNF-α, we performed cytoplasmic staining on neonatal splenocytes after Poly(A:U) stimulation. Two immune cell subsets, B220+/CD19+/MHC CII+ B cells and B220−/CD3−/CD11c−/CD11b+ myeloid cells produced detectable TNF-α in response to Poly(A:U) (Figure 6A-B). No TNF-α–producing immune cell subsets were detected in stimulated adult spleens. Lastly, to confirm that TNF-α contributed to the expansion of LIC observed in vivo, we treated neonatal Eμ-Ret mice with Poly(A:U) in the presence of TNF-α neutralizing antibody (Figure 6C). Although an isotype-matched control antibody (anti–IL-10) had no effect, neutralization of TNF-α completely blocked the Poly(A:U)-induced increase in LIC numbers. This finding demonstrates the involvement of TNF-α in the age-dependent expansion of the preleukemic cell population.
TNF-α is necessary for LIC expansion in neonates. Representative flow cytometry plots (A) and cumulative results (B) from independent experiments showing TNF-α production by neonatal and adult B220+/CD19+/MHC CII+ lymphoid cells (left) and B220–/CD11c–/CD11b+ myeloid cells (right); paired t test. (C) In vivo impact of cytokine (TNF-α or IL-10) neutralization on LIC numbers in 6-day-old Eμ-Ret pups injected with Poly(A:U). Cell counts in harvested organs were quantified using CountBright beads. (Dunnett multiple comparison test; bars represent mean ± SD). Pooled results are from at least 3 individual experiments. ∗P < .05; ∗∗P < .01, ns.
TNF-α is necessary for LIC expansion in neonates. Representative flow cytometry plots (A) and cumulative results (B) from independent experiments showing TNF-α production by neonatal and adult B220+/CD19+/MHC CII+ lymphoid cells (left) and B220–/CD11c–/CD11b+ myeloid cells (right); paired t test. (C) In vivo impact of cytokine (TNF-α or IL-10) neutralization on LIC numbers in 6-day-old Eμ-Ret pups injected with Poly(A:U). Cell counts in harvested organs were quantified using CountBright beads. (Dunnett multiple comparison test; bars represent mean ± SD). Pooled results are from at least 3 individual experiments. ∗P < .05; ∗∗P < .01, ns.
Discussion
A general mechanism to explain the apparently contradictory associations between infection and B-ALL risk reported by epidemiological studies remains elusive. In this study, we reveal that recognition of dsRNA, an intermediate or product generated during the replication of most viruses, can exert differential effects on B-ALL cells. The inclusion in our study of different B-ALL subtypes derived from both transgenic mouse models and patients demonstrates a high degree of shared response biology, at least in terms of the outcome after dsRNA exposure. This consistency across species supported the use of the Eμ-Ret model to interrogate the impact of such exposures during the preleukemic stage of disease. By targeting specific PRR, we demonstrate that both expansion and depletion of LIC can be induced by exposure to dsRNA, providing a potential mechanistic explanation for the contradictory effects attributed to early life infection.
The ability of Poly(A:U) to stimulate LIC proliferation in neonatal, but not older mice, highlights the dynamic nature of interactions between the host immune system and nascent preleukemia. Although it has been generally assumed to be the case, our study, to our knowledge, provides the first direct evidence that age-related changes in the immune response, rather than in the LIC population, influence the outcome of exposure to infection-associated stimuli. Although the complete penetrance and unpredictable latency of B-ALL in the Eμ-Ret model makes evaluation of disease progression kinetics challenging, we hypothesize that early expansion of the LIC population would increase the risk of subsequent malignant transformation, as has been reported for children diagnosed with B-ALL.47,48 It will be of great interest to determine whether early life dsRNA exposure can similarly drive B-ALL progression in other, less penetrant mouse models.49-51
Although no specific pathogen has been definitively associated with B-ALL, cytomegalovirus (CMV) has emerged as a strong candidate.52 CMV infection acquired during pregnancy or perinatally has been associated with childhood B-ALL52-54 and been shown to affect hematopoietic progenitors in cord blood.55,56 CMV triggers signaling through TLR3,57,58 raising the possibility that the potentially proleukemic activity observed in our study may be generated by early exposure to the virus. Although the strongest evidence for a role for CMV is reported for hyperdiploid B-ALL, we observed a similar magnitude of effect across a range of leukemia subtypes. However, our in vivo LIC experiments were performed exclusively with Eμ-Ret mice, which developed a leukemia characterized by the presence of chromosomal trisomies (Atre T, Farrokhi F, Fidanza M, Rever J, Guo M , Duey W , Jo S , Rolf N , Uzozie A , Baharvand F , Auer F , Hauer J , Grupp S , Eydoux P, Lange P, Seif A, Maxwell C, Reid,G.unpublished data, July 2023). It will be of considerable interest to evaluate the in vivo impact of early life PRR signaling in other transgenic models of B-ALL. Furthermore, the age-related outcome differences observed in our study may extend to fetal life.59 The lack of vertical transmission of CMV in mice has limited the assessment of such effects. Because dsRNA can cross the placenta,60 the use of PRR ligands described here may provide an alternative approach for investigating the impact of in utero immune modulation on LIC biology.
Several studies have implicated both proinflammatory and anti-inflammatory cytokines as modulators of B-ALL progression. IL-6, in combination with TNF-α and IL-1β, and transforming growth factor β have been reported to exert proleukemic activity in ETV6-RUNX1 models, whereas IL-10 inhibited leukemogenesis in a similar setting.61-63 IL-6 was also found to be a driver of mutant Pax5-driven B-ALL.64 Given that some of these studies report, as we do, consistent findings between mouse and human experimental models, it is tempting to speculate that the studies are reflecting the diversity of potential responses generated by inflammatory signaling. Previously, we have established the importance of type 1 and type 2 IFNs in modulating LIC fate, in which the neutralization or absence of IFNs leads to increased LIC burden in Eμ-RET mice.36,65 Consistent with those results, RNA sequencing indicated that there was decreased IFN-α and IFN-γ signaling in LIC after Poly(A:U) exposure. Although our results identify a contributing role for TNF-α in the context of responses to Poly(A:U), increased LIC proliferation in Eμ-Ret mice may reflect changes in influence of several cytokines.
The description of B-ALL modifiers that exert similar responses in mouse and human settings provides a potential explanation for contradictory epidemiologic data and a strategy to evaluate their ability to reduce progression of clinical disease. The application of immune modulatory strategies for the treatment, and perhaps prevention,66,67 of pediatric B-ALL will require a greater understanding of the balance between the host immune system and the abnormal B-cell populations. Our findings indicate that the outcome of immune modulation is likely to be influenced by both the nature of the infectious agent and the immune status of the child. We demonstrate that age is an important variable affecting response, but other modifiers of host immune responsiveness, whether genetic or environmental, could also influence outcome. The results of our study underscore the complexity of these interactions, and by demonstrating the potential to expand or deplete LIC by triggering different PRR pathways and subsequent immune responses, it highlight both the potential and risk of immune-related interventions.
Acknowledgments
The authors thank all participants of this research study and their families. The authors also acknowledge Arnawaz Bashir for excellent technical assistance and Amanda Lorentzian and Jenna Revers for assistance with RNA sequencing. The graphical abstract was created with BioRender.com.
This work was supported by operating grants from Canadian Institutes of Health Research (MOP-126122) (G.S.D.R.) and a grant from the US National Institutes of Health (CA214888) (M.L.C.). T.A. is the recipient of a Michael Cuccione Childhood Cancer Research doctoral scholarship and the BC Children’s Hospital graduate studentship. A.F. is a recipient of Michael Cuccione Childhood Cancer Research postdoctoral scholarship.
Authorship
Contribution: T.A. planned and performed experiments, analyzed and interpreted data, and wrote the manuscript; A.F. performed experiments and analyzed and interpreted results; S.J. performed experiments; S.S. analyzed data; J.D.-A. and M.L.C. provided critical reagents for the study; N.R. contributed to study development; G.S.D.R. conceived and designed the experiments, analyzed and interpreted data, and wrote the manuscript; and all authors have reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gregor S. D. Reid, BC Children’s Hospital Research Institute, 950 West 28th Ave, Room 3062, Vancouver, BC V5Z 4H4, Canada; e-mail: greid@bcchr.ca.
References
Author notes
The RNA sequencing data are deposited in the Gene Expression Omnibus database (accession number GSE244018).
Data are available on request from the corresponding author, Gregor S. D. Reid (greid@bcchr.ca).
The full-text version of this article contains a data supplement.



![Poly(A:U) induces LIC expansion in vivo. (A) Representative flow plots depicting early precursor B-cell populations (Hardy fractions [Fr] A, B, and C/C′) from the spleen and BM of PBS- or Poly(A:U)-treated mice. (B) FrA, FrB, and FrC/C′ and mature B cells (B cell) were quantified by flow cytometry from PBS- and Poly(A:U)-treated mice. Cell counts were quantified using CountBright beads and results pooled from at least 3 individual experiments (Dunnett multiple comparison test). (C) Representative flow cytometry plots depicting BrdU+ LIC in the spleen and BM from PBS- or Poly(A:U)-treated mice. (D) Early BCP proliferating in the BM of Poly(A:U)-treated compared with PBS-treated control mice, expressed as % BrdU+ B220+CD43+ cells. Results are pooled from 2 individual experiments (Šídák multiple comparisons test). Bars represent mean ± SD; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/22/10.1182_bloodadvances.2023010782/3/m_blooda_adv-2023-010782-gr4.jpeg?Expires=1763532124&Signature=v~4tikMiPZQ7shbYBXmbcBYkoMkzjaoQEMMeM1SQjDkOBmp2C~21nU~kCDVSdqqHI5JyZHlTwe6r8tGh3cCN-21gxOQ5vCiJHpIlAjN-lxhDuCaLPL-bBSq4SJK6Zj4NLh9PSbpW7yco~muDITYBLGSHZtOXcvBDpZCLXvNOmtS-HgUqJMkhskuo5qo1imlA8jXCdLlrtsnK2r7dzqmq4aqTdtUKFdYKPgtEiqK6rB~QSI-412Q4oxC9mRyMbVf9EXWF6Kz76pWz0S95y7w01VAW80RtrgfM9bCs45zje7pFKE9-zc14dHTnXZNy4OAWP3n19Kq9kUYB-WVElqkWpg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


