Key Points
CLL cells induce a pseudohypoxic state in surrounding T cells, contributing to T-cell dysfunction via autocrine purinergic signaling.
T-cell dysfunction by pseudohypoxia is reversible and amenable to therapeutic targeting.
Abstract
Acquired T-cell dysfunction is common in chronic B-cell malignancies. Given the strong connection between T-cell metabolism and function, we investigated metabolic alterations as the basis of T-cell dysfunction induced by malignant cells. Using B-cell malignant cell lines and human peripheral blood mononuclear cells, we first established a model that recapitulates major aspects of cancer-induced T-cell dysfunction. Cell lines derived from chronic lymphocytic leukemia (CLL) (PGA-1, CII, and Mec-1), but not from other B-cell malignancies, altered the T-cell metabolome by generating a pseudohypoxic state. T cells were retained in aerobic glycolysis and were not able to switch to oxidative phosphorylation (OXPHOS). Moreover, T cells produced immunosuppressive adenosine that negatively affected function by dampening the activation, which could be restored by the blocking of adenosine receptors. Subsequently, we uncovered a similar hypoxic-like signature in autologous T cells from primary CLL samples. Pseudohypoxia was reversible upon depletion of CLL cells ex vivo and, importantly, after the in vivo reduction of the leukemic burden with combination therapy (venetoclax and obinutuzumab), restoring T-cell function. In conclusion, we uncovered a pseudohypoxic program connected with T-cell dysfunction in CLL. Modulation of hypoxia and the purinergic pathway might contribute to therapeutic restoration of T-cell function.
Introduction
Metabolic plasticity is crucial for T-cell activation and differentiation, because immune and metabolic pathways are integrated to determine T-cell function.1 In the last years, it has become clear that altered metabolism is a hallmark of T-cell dysfunction in cancer.2 Tumor cells and immunosuppressive signals in the tumor microenvironment (TME)3 disturb crucial T-cell immunometabolic checkpoints. For example, alterations in fuel availability4 or byproducts of malignant cellular metabolism5,6 can reshape the T-cell metabolome. In other cases, coinhibitory receptors can interfere with signaling cascades that, in turn, regulate metabolism.7
In solid malignancies, another relevant immunosuppressive signal is hypoxia.8 Although tumor cells thrive in an oxygen-depleted environment, T-cell function is hindered by the hypoxic TME.9,10 Hypoxia is considered an obstacle to immunotherapy11,12 because of reduced traffic and function of tumor-infiltrating lymphocytes and chimeric antigen receptor T cells in the tumor bed.8 Because of the hypoxic environment, malignant cells in solid tumors release immunosuppressive factors, such as anti-inflammatory cytokines, byproducts, adenosine triphosphate (ATP)/adenosine, and other nucleosides.8,13 This can also occur in normoxic environments because of genetic mutations leading to the production and release of oncometabolites by tumor cells.14
In chronic B-cell malignancies, in which T-cell dysfunction also hampers immunotherapy,15 the role of oxygen sensing mechanisms is still unknown. Hypoxia is present in secondary lymphoid organs under physiological conditions and is exacerbated during tumor progression.16,17 However, most of this knowledge comes from multiple myeloma (MM) and the hypoxic bone marrow niche. In contrast, the assumption is that oxygen levels in peripheral blood are not limiting.18 Hence, the mechanisms driving T-cell dysfunction in chronic B-cell malignancies and the extent to which this phenomenon depends on active processes triggered by cancer cells (eg, hypoxia and oncometabolites production) have remained obscure.
Previously, we described aspects of T-cell dysfunction in primary chronic lymphocytic leukemia (CLL).19 However, substantial interpatient variation complicates investigation of general underlying principles. Therefore, to dissect the metabolic interplay between malignant B cells and T cells, we first established an in vitro coculture model that recapitulates major aspects of cancer-induced T-cell dysfunction observed in patients.15 We found that leukemic cells of CLL origin, specifically, were able to alter T-cell metabolism and oxygen sensing, inducing a hypoxic-like program in aerobic conditions and suppression of T-cell function. Importantly, primary CLL cells similarly induced a pseudohypoxic signature in autologous T cells. Upon removal of CLL cells, either ex vivo or with venetoclax + obinutuzumab treatment in vivo, the expression of hypoxic genes was normalized, and T-cell function was recovered.
Methods
HD PBMCs and malignant cell lines coculture assay
Peripheral blood mononuclear cells (PBMCs) from healthy donors (HDs) (supplemental Table 3) were derived from buffycoats obtained from Sanquin (Amsterdam, The Netherlands) and cultured in the presence or absence of cancer cell lines, namely PGA-1, CII and Mec-1, LME-1, U-266 and RPMI-8226, and Nalm-6 (supplemental Table 8). T cells were stimulated with anti-CD3 (clone 1XE) and anti-CD28 (clone 15E8) soluble antibodies and analyzed 24 and 48 hours after stimulation, as indicated. Treatments applied to the coculture are described in detail in the supplemental Data.
Patient material
Peripheral blood was obtained from patients with CLL and age-matched HDs (supplemental Table 3) after obtaining written and informed consent in accordance with the Declaration of Helsinki. PBMCs from HDs and patients with CLL were analyzed at baseline or cultured in RPMI supplemented with 10% fetal calf serum and stimulated with anti-CD3/CD28 soluble antibodies for 48 hours. T cells from patients with CLL were also analyzed upon depletion of CLL cells using an EasySep Human T Cell Enrichment Kit (supplemental Table 4). In addition, PBMCs were collected from patients with CLL enrolled in the phase 2 HOVON139 trial20 (Netherlands Trial Register ID NTR6043; supplemental Table 3) at baseline and after 12 to 24 treatment cycles. Treatment consisted of 2 preinduction cycles with obinutuzumab (3000 mg in cycle 1 and 1000 mg in cycle 2), followed by combinations of venetoclax (augmented to 400 mg) and obinutuzumab (1000 mg; cycles 3-8), and venetoclax monotherapy (400 mg; cycles 9-14). Ethical approval was provided by the medical ethical committee at the Amsterdam University Medical Centers, location AMC, Amsterdam.
Flow cytometry
Cells were stained with cell dyes or monoclonal antibodies as described previously.19 Detailed description is provided in supplemental Files.
Metabolic analyses
Extracellular flux analysis and intracellular metabolomics were performed on isolated CD4+ T cells after the coculture experiment. Adenosine and purines measurements were performed using supernatants. More detailed explanations can be found in supplemental Files.
Transcriptomics
Transcriptomic analysis was performed using isolated CD4+ T cells after the coculture experiment or after a 48-hour stimulation of CLL T cells in the presence or absence of the autologous CLL cells. More details are reported in supplemental Files.
Real-time qPCR
Gene expression analysis by real-time quantitative polymerase chain reaction (qPCR) was performed using standard techniques, explained in detail in supplemental Files. Probes are referenced in supplemental Table 6.
Data presentation and statistical analysis
Data are presented as mean ± standard error of the mean. Data were analyzed using paired t test (when comparing the mean of 2 groups) or 1-way analysis of variance followed by Tukey multiple comparison test (when comparing the mean of >2 groups), as indicated in figure legends. Statistical tests were performed using GraphPad Prism version 9.
Results
T-cell function is dampened in parallel to deregulated metabolism by malignant B cells, especially from CLL origin
We evaluated whether malignant B-cell lines from a distinct origin induce T-cell dysfunction. PBMCs from HDs were stimulated with anti-CD3/28 antibodies in the presence or absence of PGA-1, LME-1, and Nalm-6 cells derived from CLL, MM, and acute lymphoblastic leukemia, respectively (supplemental Figure 1A). Proliferation of CD4+ (Figure 1A; supplemental Figure 1B) and CD8+ T cells (supplemental Figure 1C) was dampened by the presence of malignant cells. PGA-1 cells abolished CD4+ T-cell proliferation, whereas LME-1 and Nalm-6 cells did not completely abrogate it. Furthermore, PGA-1 cells impaired the production of tumor necrosis factor α, interleukin-2, and interferon-gamma by CD4+ (and CD8+, data not shown) T cells. The membrane localization of the degranulation marker CD107A was unaffected (Figure 1B). Cytokine production by T cells was preserved in the presence of LME-1 or Nalm-6 cells, indicating that PGA-1 induces a distinct type of T-cell dysfunction that also encompasses reduced effector function. Importantly, coculture with cell lines did not decrease viability of the T cells (supplemental Figure 1D).
T-cell function is dampened in parallel to deregulated metabolism in the presence of malignant B cells, especially from CLL origin. PBMCs from HDs were thawed and cultured in the presence or absence of PGA-1, CII, and Mec-1 derived from CLL; LME-1, U-266, and RPMI-8226 derived from MM; and Nalm-6 cells derived from acute B-lymphoblastic leukemia. After 24 hours, anti-CD3/CD28 antibodies were added, and T cells were analyzed 48 hours after stimulation. (A) PBMCs were stained with carboxyfluorescein succinimidyl ester, and CD4+ T-cell proliferation was assessed. (B) Brefeldin A and Golgi Stop were added to the coculture 4 hours before analysis to assess cytokine production and degranulation. Percentage of CD4+ T cells producing tumor necrosis factor (TNF)-α, interleukin-2 (IL-2), and interferon-gamma (IFNγ) 48 hours after stimulation and percentage of cells expressing CD107A at the cell surface is plotted. After gating on CD4+ T cells, activated T cells (CD4+CD25+) were analyzed for (C) T-cell activation, represented by CD25 levels (gMFI); metabolic capacity, as indicated by (D) glucose uptake (2-NBDG); and (E) mitochondrial mass (MitoTracker green FM). (F) Activated T cells were analyzed for the expression of CD25 and (G) effector function after coculture with CII, Mec-1, U-266, and RPMI-8226. Data are presented as mean ± standard error of the mean (SEM). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05; ns, not significant (1-way analysis of variance [ANOVA] followed by Tukey multiple comparison test or paired t test).
T-cell function is dampened in parallel to deregulated metabolism in the presence of malignant B cells, especially from CLL origin. PBMCs from HDs were thawed and cultured in the presence or absence of PGA-1, CII, and Mec-1 derived from CLL; LME-1, U-266, and RPMI-8226 derived from MM; and Nalm-6 cells derived from acute B-lymphoblastic leukemia. After 24 hours, anti-CD3/CD28 antibodies were added, and T cells were analyzed 48 hours after stimulation. (A) PBMCs were stained with carboxyfluorescein succinimidyl ester, and CD4+ T-cell proliferation was assessed. (B) Brefeldin A and Golgi Stop were added to the coculture 4 hours before analysis to assess cytokine production and degranulation. Percentage of CD4+ T cells producing tumor necrosis factor (TNF)-α, interleukin-2 (IL-2), and interferon-gamma (IFNγ) 48 hours after stimulation and percentage of cells expressing CD107A at the cell surface is plotted. After gating on CD4+ T cells, activated T cells (CD4+CD25+) were analyzed for (C) T-cell activation, represented by CD25 levels (gMFI); metabolic capacity, as indicated by (D) glucose uptake (2-NBDG); and (E) mitochondrial mass (MitoTracker green FM). (F) Activated T cells were analyzed for the expression of CD25 and (G) effector function after coculture with CII, Mec-1, U-266, and RPMI-8226. Data are presented as mean ± standard error of the mean (SEM). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05; ns, not significant (1-way analysis of variance [ANOVA] followed by Tukey multiple comparison test or paired t test).
Upon exposure to PGA-1 but not LME-1 or Nalm-6 cells, activated CD4+ and CD8+ T cells showed reduced expression of activation markers CD25 (interleukin-2 receptor) and CD71 (transferrin receptor-1; Figure 1C; supplemental Figure 1E-H). Glucose uptake and mitochondrial mass, key parameters for T-cell function, were evaluated using 2-NBDG and MitoTracker green FM, respectively, and were found decreased only in the presence of PGA-1 cells (Figure 1D-E; supplemental Figure 1H). To assess whether other cell types present in the PBMC fraction were involved in the induction of T-cell dysfunction, either isolated CD3 T cells or the whole PBMC fraction were cocultured with PGA-1. Clearly, PGA-1 induced T-cell dysfunction in both setups, indicating that PGA cells induce this phenotype in a direct manner (supplemental Figure 2A).
Given that cancer cells can limit nutrient availability in the TME,6 competition for glucose might cause failure in upregulating the CD25 expression on the T cells. Concurrently, we explored the potential role of cancer cell waste product lactate in altering T-cell function.4,21 Glucose or lactate supplementation to HD T cells did not alleviate or induce, respectively, the T-cell dysfunction in this model (supplemental Figure 2B-C). Furthermore, conditioned media from PGA-1 cells did not affect CD25 expression or glucose uptake, and the use of a transwell system rescued both glucose uptake and CD25 expression (supplemental Figure 2D). These results demonstrate that PGA-1 induce T-cell dysfunction upon close interaction and that neither the lack of glucose4 nor the presence of tumor-derived lactate21 are responsible for the suppressed T-cell activation. To corroborate these findings, more cell lines were investigated, including CII and Mec-1 deriving from CLL and U-266 and RPMI-8226 of MM origin (supplemental Table 8). T-cell function was dampened in the presence of CII and Mec-1 (CD25 expression and cytokine production; Figure 1F-G); exposure to U-266 did not affect T-cell function, whereas RPMI-8226 cells boosted T-cell cytokine production. Proliferation capacity of the cell lines showed no correlation with the induction of T-cell dysfunction (supplemental Figure 1I). Together, these results indicated that cell lines of CLL origin are able to dampen T-cell activation.
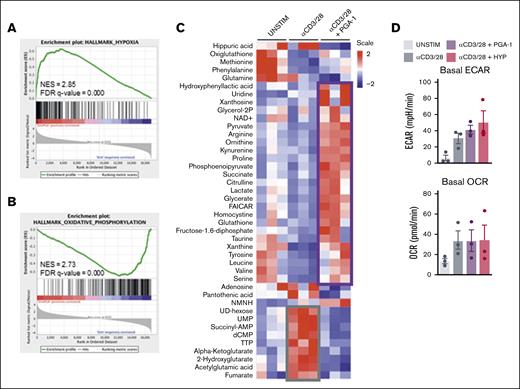
PGA-1 cells induce a hypoxic gene signature and metabolic alterations in T cells, skewing the fate of pyruvate into aerobic glycolysis
To investigate this specific defect in T-cell activation more thoroughly, we first applied RNA sequencing analysis, which revealed a profoundly different transcriptomic profile in CD4+ T cells stimulated in the presence of PGA-1 cells (supplemental Figure 3A). Gene set enrichment analysis (GSEA) against Hallmark Signatures (h.all.v7.4) identified hypoxia as the most significantly upregulated pathway in T cells activated in the presence of PGA-1 (Figure 2A; supplemental Figure 3B), as shown by the higher expression of genes promoting glycolysis (HK2, PDK1, SLC2A3, and ENO2), angiogenesis (VEGFA) and apoptosis (BNIP3L and BCL2), or dampening mitochondrial biogenesis (KDM3A) (supplemental Figure 2B-C). Mitochondrial processes such as OXPHOS (Figure 2B) and fatty acid β-oxidation as well as the expression of MYC targets were downregulated (supplemental Figure 3B). The lists of differentially expressed genes within the gene sets hypoxia and OXPHOS are available in supplemental Tables 1 and 2. GSEA also suggested the induction of an inflammatory response during stimulation in the presence of PGA-1 (supplemental Figure 3B).
PGA-1 cells induce a hypoxic gene signature and profound metabolic alterations in T cells, skewing the fate of pyruvate into aerobic glycolysis. PBMCs were cultured with and without PGA-1 cells and stimulated with αCD3/CD28. After stimulation, CD4+ T cells were sorted and lysed for RNA sequencing (48 hours after stimulation), metabolomics (48 hours after stimulation), and extracellular flux analysis (24 hours after stimulation). (A-B) Enrichment plot showing hypoxic gene signature and downregulation of OXPHOS in CD4+ T cells exposed to PGA-1. On the x-axis, red indicates the presence of PGA-1 cells, whereas blue represents PBMC stimulated alone. Curves (green) indicate cumulative enrichment quantified by enrichment score on the y-axis. Tick marks on the x-axis correspond to the rank of genes in the gene set. (C) Heat map representing the abundance of metabolites in CD4+ T cells unstimulated and stimulated in presence or absence of PGA-1. Metabolomics data were analyzed with Tercen software and scaled and hierarchical clustered by rows and columns, and significantly different metabolites in T cells stimulated in the presence of PGA-1 compared with the stimulated control (paired t test) were plotted. Metabolites abundance in unstimulated cells were plotted as a comparison. Two main clusters were identified: metabolites increasing after T-cell stimulation (gray box) and metabolites increasing when T cells are stimulated in the presence of PGA-1 cells (purple box). (D) Extracellular flux analysis showing extracellular acidification rate and oxygen consumption rate of sorted unstimulated CD4+ T cells, stimulated for 24 hours in the presence or absence of PGA-1 and stimulated in hypoxia (HYP; 1% O2) as a positive control. FDR, false discovery rate; NES, normalized enrichment score; UNSTIM, unstimulated.
PGA-1 cells induce a hypoxic gene signature and profound metabolic alterations in T cells, skewing the fate of pyruvate into aerobic glycolysis. PBMCs were cultured with and without PGA-1 cells and stimulated with αCD3/CD28. After stimulation, CD4+ T cells were sorted and lysed for RNA sequencing (48 hours after stimulation), metabolomics (48 hours after stimulation), and extracellular flux analysis (24 hours after stimulation). (A-B) Enrichment plot showing hypoxic gene signature and downregulation of OXPHOS in CD4+ T cells exposed to PGA-1. On the x-axis, red indicates the presence of PGA-1 cells, whereas blue represents PBMC stimulated alone. Curves (green) indicate cumulative enrichment quantified by enrichment score on the y-axis. Tick marks on the x-axis correspond to the rank of genes in the gene set. (C) Heat map representing the abundance of metabolites in CD4+ T cells unstimulated and stimulated in presence or absence of PGA-1. Metabolomics data were analyzed with Tercen software and scaled and hierarchical clustered by rows and columns, and significantly different metabolites in T cells stimulated in the presence of PGA-1 compared with the stimulated control (paired t test) were plotted. Metabolites abundance in unstimulated cells were plotted as a comparison. Two main clusters were identified: metabolites increasing after T-cell stimulation (gray box) and metabolites increasing when T cells are stimulated in the presence of PGA-1 cells (purple box). (D) Extracellular flux analysis showing extracellular acidification rate and oxygen consumption rate of sorted unstimulated CD4+ T cells, stimulated for 24 hours in the presence or absence of PGA-1 and stimulated in hypoxia (HYP; 1% O2) as a positive control. FDR, false discovery rate; NES, normalized enrichment score; UNSTIM, unstimulated.
Secondly, the alterations induced by PGA-1 cells on T-cell metabolism were investigated by liquid chromatography-mass spectrometry (Figure 2C). Metabolites enhanced during stimulation in the presence of PGA-1 (purple box) were distinct compared with those increased upon stimulation alone (gray box). Enrichment analysis to assess which metabolic pathways were induced upon T-cell stimulation and which were affected by the presence of PGA-1 showed that stimulated T cells upregulated both anabolic and catabolic pathways (supplemental Figure 3D). In contrast, when T cells were stimulated in the presence of PGA-1, glycolysis and gluconeogenesis were increased, whereas the processes taking place in the mitochondria were down-represented (supplemental Figure 3E). Accordingly, we observed the accumulation of glycolytic intermediates such as pyruvate, phosphoenolpyruvate, and lactate in the presence of PGA-1 (Figure 2C). These data, together with a general reduction in the intermediates of the Krebs cycle, indicated that PGA-1 cells skewed the utilization of glucose by T cells in aerobic glycolysis rather than in OXPHOS.
To study the effect of a hypoxic gene signature, healthy T cells were stimulated in the presence or absence of PGA-1 cells, sorted using fluorescence-activated cell sorter, and analyzed by extracellular flux analysis. T cells stimulated in hypoxic conditions (1% O2) were taken along as positive control. T cells activated in the presence of PGA-1 cells or in hypoxia exhibited higher basal extracellular acidification rate, consistent with an hypoxic response. Because the flux analyses were performed after reoxygenation, the oxygen consumption rate was unchanged (Figure 2D; supplemental Figure 3F).
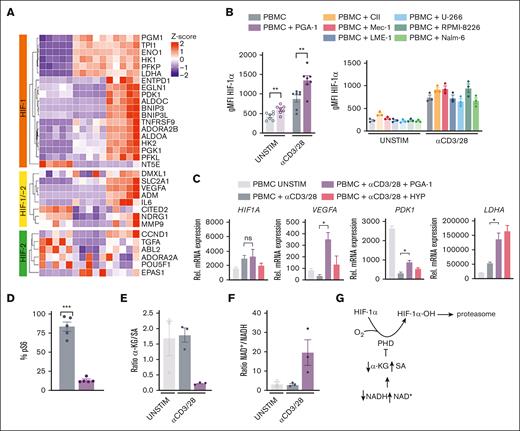
PGA-1 induced hypoxic signature in T cells is due to pseudohypoxia, which correlates with HIF-1α stabilization and expression of its target genes
Adaptation to low oxygen levels is mediated by the family of hypoxia-inducible factors (HIFs),22 which comprises 2 α isoforms, HIF-1α (HIF1A) and HIF-2α (EPAS1), and 1 β isoform, HIF-1β (ARNT). The half-life of the α subunits is tightly regulated by oxygen levels through the activity of prolyl hydroxylases. In hypoxia, HIF-α is stabilized and can induce an array of target genes promoting metabolic and functional adaptations to low oxygen levels. Importantly, although there was no difference in the expression of HIF1A, EPAS1, or ARNT, the expression of HIF-1 target genes and HIF-1/-2 common targets was upregulated in T cells exposed to PGA-1, whereas HIF-2 targets were not affected (Figure 3A). Furthermore, HIF-1α protein levels were higher in T cells stimulated in the presence of PGA-1 (Figure 3B). Of note, a small increase in HIF-1α was observed upon stimulation in the absence of PGA-1, because this transcription factor plays a role in the glycolytic switch upon T-cell receptor (TCR) triggering23,24 (Figure 3A-B). Increased expression of HIF-1α was also observed in T cells after contact with CII, Mec-1 cells, and RPMI-8226 (Figure 3B). In the presence of the CLL cell lines (CII and Mec-1), the increase in HIF-1α correlated with reduced T-cell activation and impaired function. Instead, accumulation of this transcription factor after contact with RPMI-8226 cells reflected the enhanced T-cell activation status induced by contact with those cells, reflecting the duality of the role of HIF-1α/hypoxia in T-cell biology.23,25
PGA-1 induced hypoxic signature in T cells is due to pseudohypoxia and correlates with HIF-1α stabilization and expression of its target genes. (A) PBMCs were thawed and cultured with and without PGA-1 cells and stimulated with anti-CD3/28. 48 hours after stimulation, CD4+ T cells were sorted and lysed for RNA sequencing and purified using magnetic beads for intracellular metabolite analysis. Heat map showing the expression of HIF-1, HIF-2, and common targets. (B) HIF-1α assessment by flow cytometry 24 hours after stimulation, when the expression of CD25 in activated T cells is comparable between stimulation in the presence or absence of cell lines. (C) Gene expression of HIF1A, VEGFA, PDK1, and LDHA was evaluated by qPCR at 24 hours to confirm RNA sequencing data. In parallel, PBMCs were stimulated with anti-CD3/28 in hypoxia (HYP, PO2 = 1%), as a control. (D) Fraction of pS6+ T cells was evaluated by flow cytometry. (E) Intracellular α-KG / SA (arbitrary units, assuming equal ionization efficiency of α-KG and SA, according to similarity in their structure) in CD4+ T cells stimulated in the presence or absence of PGA-1. (F) Intracellular NAD+ / NADH ratio represented in arbitrary units. (G) Regulation of HIF-1α stability by oxygen and pseudohypoxia. Data are presented as mean ± SEM. ∗∗∗P < .001; ∗P < .05; ns (1-way ANOVA followed by Tukey multiple comparison test). α-KG / SA, α-ketoglutarate–to–succinate ratio; ns, not significant; PHD, prolyl hydroxylase. Scheme was generated with BioRender.com.
PGA-1 induced hypoxic signature in T cells is due to pseudohypoxia and correlates with HIF-1α stabilization and expression of its target genes. (A) PBMCs were thawed and cultured with and without PGA-1 cells and stimulated with anti-CD3/28. 48 hours after stimulation, CD4+ T cells were sorted and lysed for RNA sequencing and purified using magnetic beads for intracellular metabolite analysis. Heat map showing the expression of HIF-1, HIF-2, and common targets. (B) HIF-1α assessment by flow cytometry 24 hours after stimulation, when the expression of CD25 in activated T cells is comparable between stimulation in the presence or absence of cell lines. (C) Gene expression of HIF1A, VEGFA, PDK1, and LDHA was evaluated by qPCR at 24 hours to confirm RNA sequencing data. In parallel, PBMCs were stimulated with anti-CD3/28 in hypoxia (HYP, PO2 = 1%), as a control. (D) Fraction of pS6+ T cells was evaluated by flow cytometry. (E) Intracellular α-KG / SA (arbitrary units, assuming equal ionization efficiency of α-KG and SA, according to similarity in their structure) in CD4+ T cells stimulated in the presence or absence of PGA-1. (F) Intracellular NAD+ / NADH ratio represented in arbitrary units. (G) Regulation of HIF-1α stability by oxygen and pseudohypoxia. Data are presented as mean ± SEM. ∗∗∗P < .001; ∗P < .05; ns (1-way ANOVA followed by Tukey multiple comparison test). α-KG / SA, α-ketoglutarate–to–succinate ratio; ns, not significant; PHD, prolyl hydroxylase. Scheme was generated with BioRender.com.
HIF-1 targets such as VEGF, LDHA, and PDK1 were already increased 24 hours after stimulation in the presence of PGA-1, whereas HIF1A expression itself was not affected, as confirmed by qPCR (Figure 3C), demonstrating that HIF-1α activity was higher and shaped the transcriptome accordingly. HIF-1α protein synthesis can be regulated in an oxygen-independent way through mTOR.24 However, the phosphorylation of S6 (p-S6) was lower in T cells stimulated in the presence of PGA-1 (Figure 3D), indicating reduced mTOR signaling and arguing against this mechanism for HIF-1α stabilization in this model. Because of the fact that the coculture experiments were conducted in normoxia, we hypothesized that HIF-1α was stabilized at a posttranslational level in an oxygen-independent manner. We observed a reduced α-ketoglutarate–to–succinate ratio in T cells exposed to PGA-1 (Figure 3E), together with an increased cytosolic ratio of free NAD+ to reduced NAD (Figure 3F), which are markers of pseudohypoxia,26,27that is, the inhibition of HIF-1α hydroxylation and degradation in normoxic conditions through metabolic imbalance (Figure 3G).
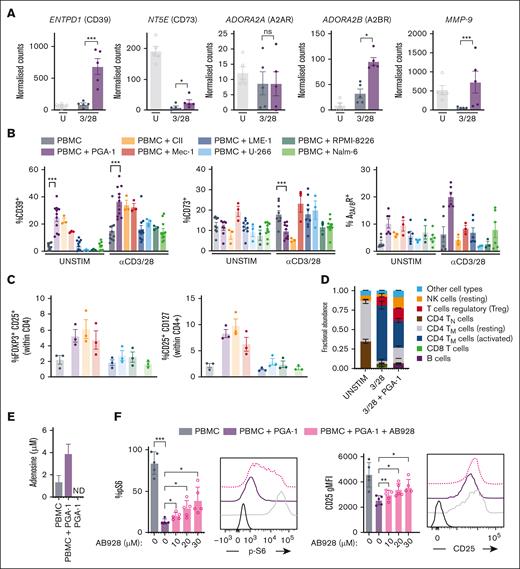
Pseudohypoxia induces extracellular adenosine production and purinergic signaling in an autocrine manner, thereby inducing T-cell dysfunction
The data collected so far indicated that PGA-1–induced T-cell dysfunction is caused by sustained glycolytic activity because of pseudohypoxia, which, in turn, alters T-cell oxygen sensing and gene transcription. Besides genes involved in glycolysis, cell cycle arrest, and apoptosis, the purinergic machinery, which can drive immunosuppression,28 was upregulated in T cells exposed to PGA-1 cells. ENTPD1 (CD39), NT5E (CD73), MMP9 (matrix metalloproteinase 9), and ADORA2B (A2BR) were higher expressed in T cells activated in the presence of PGA-1, whereas ADORA2A (A2AR) was not (Figure 4A). Importantly, ENTPD1, NT5E, ADORA2B, and MMP-9 are known HIF1-α transcriptional targets29 (Figure 3A).
Pseudohypoxia induces extracellular adenosine production and purinergic signaling in an autocrine manner, thereby inducing T-cell dysfunction. (A) Normalized counts indicating the expression of genes ENTPD1 (CD39), NT5E (CD73), ADORA2A (A2AR), ADORA2B (A2BR), and MMP-9. (B) PBMCs from HDs were thawed and cultured in the presence or absence of PGA-1, CII, Mec-1, LME-1, U-266, RPMI-8226, and Nalm-6 cells and stimulated with anti-CD3/CD28 antibodies. CD4+ T cells were analyzed 48 hours after stimulation by multiparameter flow cytometry for the expression of CD39, CD73, and A2A/BR expression on the cell surface. (C) Percentage of FOXP3+CD25+ and CD25+CD127– cells within the CD4 compartment after 72 hours coculture with the cell lines, in absence of stimulation. (D) Deconvolution of RNA sequencing data with CIBERSORTx analysis; fractional abundance of indicated immune cell subsets is plotted. (E) Supernatants from the coculture stimulated with anti-CD3/CD28 antibodies were analyzed for adenosine concentration; (F) p-S6 and CD25 expression was assessed in presence of the A2A/BR antagonist AB928 (10-30 μM). Panel D represents the same data points as that of Figure 3D in absence of AB928. Data are presented as mean ± SEM. ∗∗∗P < .001; ∗∗P < .01; ∗P < .05; ns (1-way ANOVA followed by Tukey multiple comparison test). ND, not detected; U, unstim.
Pseudohypoxia induces extracellular adenosine production and purinergic signaling in an autocrine manner, thereby inducing T-cell dysfunction. (A) Normalized counts indicating the expression of genes ENTPD1 (CD39), NT5E (CD73), ADORA2A (A2AR), ADORA2B (A2BR), and MMP-9. (B) PBMCs from HDs were thawed and cultured in the presence or absence of PGA-1, CII, Mec-1, LME-1, U-266, RPMI-8226, and Nalm-6 cells and stimulated with anti-CD3/CD28 antibodies. CD4+ T cells were analyzed 48 hours after stimulation by multiparameter flow cytometry for the expression of CD39, CD73, and A2A/BR expression on the cell surface. (C) Percentage of FOXP3+CD25+ and CD25+CD127– cells within the CD4 compartment after 72 hours coculture with the cell lines, in absence of stimulation. (D) Deconvolution of RNA sequencing data with CIBERSORTx analysis; fractional abundance of indicated immune cell subsets is plotted. (E) Supernatants from the coculture stimulated with anti-CD3/CD28 antibodies were analyzed for adenosine concentration; (F) p-S6 and CD25 expression was assessed in presence of the A2A/BR antagonist AB928 (10-30 μM). Panel D represents the same data points as that of Figure 3D in absence of AB928. Data are presented as mean ± SEM. ∗∗∗P < .001; ∗∗P < .01; ∗P < .05; ns (1-way ANOVA followed by Tukey multiple comparison test). ND, not detected; U, unstim.
Flow cytometry confirmed the increased expression of CD39 at the cell surface when T cells were exposed to PGA-1, CII, and Mec-1 from CLL origin. In contrast, CD39 on the T-cell surface was unaffected in the presence of LME-1, U-266, RPMI-8226, or Nalm-6 cells (Figure 4B). CD73 as well as A2A/BR expression was rather variable, as previously shown.17 Of note, CD73 was still perturbed by the presence of CLL cell lines, specifically. Although induced transcriptionally in the presence of PGA-1, the protein level of CD73 at the cell surface was decreased, and this perturbation was caused by PGA-1 and CII cell lines, whereas Mec-1 induced the upregulation of CD73 on the T-cell surface. The higher expression of MMP-9, which is the metalloprotease processing the shedding of CD73 from the plasma membrane, might indicate that CD73 is present in a soluble form.
Because CD39 and CD73 are expressed by T regulatory (Treg) cells,30,31 we investigated whether contact with the cell lines would induce (Treg) cell formation in a nonstimulated setting. CLL-derived cell lines (PGA-1, CII, and Mec-1) resulted in a higher percentage of FOXP3+CD25+ or CD25+CD127– T cells in the CD4 compartment (Figure 4C). Moreover, we assessed the abundance of immune cell population in an unbiased manner, by applying the CIBERSORTx approach.32 Deconvolution of our RNA sequencing data set also confirmed that stimulation in the presence of PGA-1 induced Treg cell formation (Figure 4D).
Because CD39 and CD73 mediate the conversion of ATP to the immunosuppressive metabolite adenosine, we checked adenosine levels. Adenosine accumulated in the supernatant when T cells were activated in the presence of PGA-1 but was not produced by PGA-1 alone, indicating it was produced by pseudohypoxic T cells (Figure 4E). To corroborate this finding, expression of purinergic markers by the various cell lines was investigated (supplemental Figure 4A). In general, all the cell lines expressed CD39, with PGA-1, CII, and Mec-1 showing the highest level. CD73 was barely expressed by all the cell lines, with the exception of Mec-1, and the A2A/BR was variable. Given the expression of the machinery for adenosine production on PGA-1, CII, and Mec-1, we concluded that malignant cells might contribute to adenosine production in the coculture, in the presence of the substrate. In addition, the cell lines are not affected by the presence of T cells regarding the expression of purinergic markers (supplemental Figure 4B). Moreover, targeted metabolomics analysis of the supernatants from T cells exposed to PGA-1 showed extracellular accumulation of other purines and their precursors, such as AICAR, guanine, guanosine, hypoxanthine, and pseudouridine (supplemental Figure 4C-D). Specifically, hypoxanthine and pseudouridine are considered markers of hypoxic stress.33
In order to assess whether adenosine production was contributing to PGA-1–induced T-cell dysfunction, we used the A2A/BR antagonist Etrumadent/AB928. Supplementation of AB928 during stimulation in the presence of PGA-1 cells partially restored TCR signaling, as shown by increased pS6 and expression of CD25 (Figure 4F; supplemental Figure 4E).
Primary CLL cells induce a hypoxic signature on autologous T cells
It has been previously shown that primary CLL cells directly impair the function of autologous T cells in terms of reduced activation, proliferation, cytotoxicity, an increased regulatory phenotype, and expression of inhibitory receptors.34,35 The transcriptomic profile of T cells stimulated in the presence or absence of autologous CLL cells36 was compared with that of HD T cells exposed to PGA-1 cells. GSEA revealed hypoxia as well as downregulation of OXPHOS as one of the common enriched pathways in both data sets (Figure 5A). Intriguingly, metabolic alterations indicating pseudohypoxia were also found in T cells from patients with CLL when compared with T cells from HDs, as shown by reduced α-ketoglutarate–to–succinate ratio (Figure 5B) and increased NAD+–to–reduced NAD ratio (Figure 5C).
Primary CLL cells induce a hypoxic signature on autologous T cells. (A) CLL cells were depleted from their PBMCs fraction using the EasySep T cell enrichment kit. The whole PBMC fraction (CLL) or the isolated fraction were stimulated with anti-CD3/28 antibodies for 48 hours. CD4+ T cells were sorted using fluorescence-activated cell sorter for transcriptomic analysis and GSEA was performed on normalized counts (DESeq2 median of ratios normalization) using Hallmark Signatures (h.all.v7.4). This analysis was compared with GSEA performed on HD T cells cultured in the presence or absence of PGA-1 cells, and common altered pathway are shown in the dotplot. Color scale indicate the mean NES of Hallmark pathways, with NES > 0 representing upregulation of specified pathway in CD4+ T cells stimulated in the presence of CLL cells or PGA-1 and NES < 0 denoting downregulation of the specified pathways. Significance is represented by the negative value of log10q (minlog10q) and expressed by the size of the dots. (B) Plot representing the ratio of intracellular α-KG to SA in arbitrary units. (C) Plot representing the ratio of NAD+ to reduced NAD in arbitrary units. (D) PBMC from patients with CLL and HDs were thawed and stimulated with anti-CD3/28 antibodies and CD4+ T cells were analyzed for CD39, CD73, and A2A/BR expression. (E) CLL cells were depleted from PBMCs from patients with CLL with EasySep T cell enrichment kit. The whole PBMC fraction (CLL) or the isolated fraction (iso) were stimulated with anti-CD3/28 antibodies for 48-hours, and CD4+ T cells were analyzed by flow cytometry for expression of CD39 and HIF-1α. (F) PBMCs of patients with CLL before and after undergoing 12 to 24 cycles of venetoclax plus obinutuzumab were analyzed by flow cytometry for expression of CD39, CD73, A2A/BR, and HIF1 on CD4+ T cells after a 48-hour stimulation with anti-CD3/CD28 antibodies. Data are presented as mean ± SEM. ∗∗∗P < .001; ∗∗P < .01, ∗P < .05; ns (1-way ANOVA followed by Tukey multiple comparison test or t test).
Primary CLL cells induce a hypoxic signature on autologous T cells. (A) CLL cells were depleted from their PBMCs fraction using the EasySep T cell enrichment kit. The whole PBMC fraction (CLL) or the isolated fraction were stimulated with anti-CD3/28 antibodies for 48 hours. CD4+ T cells were sorted using fluorescence-activated cell sorter for transcriptomic analysis and GSEA was performed on normalized counts (DESeq2 median of ratios normalization) using Hallmark Signatures (h.all.v7.4). This analysis was compared with GSEA performed on HD T cells cultured in the presence or absence of PGA-1 cells, and common altered pathway are shown in the dotplot. Color scale indicate the mean NES of Hallmark pathways, with NES > 0 representing upregulation of specified pathway in CD4+ T cells stimulated in the presence of CLL cells or PGA-1 and NES < 0 denoting downregulation of the specified pathways. Significance is represented by the negative value of log10q (minlog10q) and expressed by the size of the dots. (B) Plot representing the ratio of intracellular α-KG to SA in arbitrary units. (C) Plot representing the ratio of NAD+ to reduced NAD in arbitrary units. (D) PBMC from patients with CLL and HDs were thawed and stimulated with anti-CD3/28 antibodies and CD4+ T cells were analyzed for CD39, CD73, and A2A/BR expression. (E) CLL cells were depleted from PBMCs from patients with CLL with EasySep T cell enrichment kit. The whole PBMC fraction (CLL) or the isolated fraction (iso) were stimulated with anti-CD3/28 antibodies for 48-hours, and CD4+ T cells were analyzed by flow cytometry for expression of CD39 and HIF-1α. (F) PBMCs of patients with CLL before and after undergoing 12 to 24 cycles of venetoclax plus obinutuzumab were analyzed by flow cytometry for expression of CD39, CD73, A2A/BR, and HIF1 on CD4+ T cells after a 48-hour stimulation with anti-CD3/CD28 antibodies. Data are presented as mean ± SEM. ∗∗∗P < .001; ∗∗P < .01, ∗P < .05; ns (1-way ANOVA followed by Tukey multiple comparison test or t test).
To assess whether the increased hypoxic gene signature in T cells from patients with CLL was correlated with altered purinergic signaling, we compared the expression of enzymes involved in adenosine production and sensing. Reduced activation of CLL T cells (supplemental Figure 5A) was parallel to increased CD39 expression, compared with that of HD cells, both in unstimulated and stimulated conditions, with no alterations in CD73 expression (Figure 5D; supplemental Figure 5B). Contrary to what we observed in T cells cocultured with PGA-1, levels of A2A/BR were higher in T cells from HDs (Figure 5D). In order to assess the contribution of CLL cells to the observed phenotype, T-cell stimulation was performed in isolated T cells from CLL PBMCs upon CLL depletion ex vivo. Depletion of CLL cells restored activation and the expression of CD39 and HIF-1α to the levels of HD (supplemental Figure 5C-D; Figure 5E). HIF-1α followed the same trend as CD25, mirroring the activation status of the T cell.37 CLL cells expressed CD39 to a higher extent than HD B cells, exhibiting a larger double positive (CD39+CD73+) population, possibly contributing to adenosine production together with autologous T cells (supplemental Figure 5E).
Lastly, we examined whether in vivo CLL cells depletion similarly affects the level of CD39 and adenosine receptors. Samples from 6 patients enrolled in the randomized phase 2 HOVON139 trial20 were analyzed at baseline (pre) and after treatment with venetoclax and obinutuzumab (post) (see supplemental Table 3 for patient characteristics). T cells from posttreatment samples showed improved T-cell activation, as indicated by increased CD25 and HIF-1α, and decreased CD39, with no major differences in A2A/BR expression when compared with posttreatment samples (Figure 5F; supplemental Figure 5G). Thus, elimination of CLL cells, either ex vivo or in vivo, prevents the hypoxic gene signature and expression of CD39 characterizing dysfunctional CLL T cells, indicating an important role for this pathway in T-cell (dys)function in CLL.
Discussion
In this study, we have shown that CLL-derived cell lines induce a hypoxic-like program in T cells, independently of oxygen levels. This phenotype was not induced by cell lines derived from MM and acute lymphoblastic leukemia. Importantly, hypoxic features were recapitulated in T cells from patients with CLL, suggesting that this way of disturbing T-cell immunity is specific to leukemic cells of CLL origin. Because leukemia is predominantly a liquid tumor, the role of hypoxic signaling in dampening T-cell immunity in peripheral blood was presumed to be irrelevant.16 Furthermore, T-cell hypoxia as well as CD39 expression was reversible upon reduction of the leukemic burden with venetoclax + obinutuzumab.
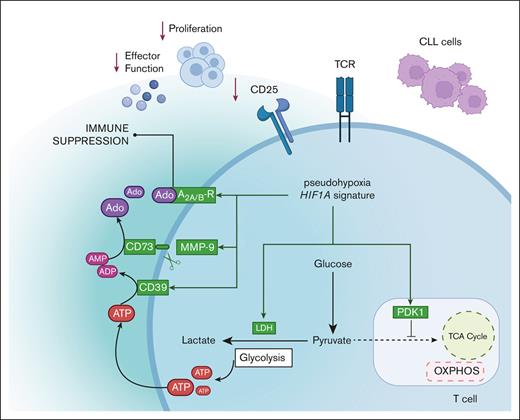
In the presence of PGA-1 cells, the acquisition of an intracellular pseudohypoxic environment, caused by alterations of T-cell metabolome, dampened T-cell immunity by 2 mechanisms. First, T cells were not able to switch to OXPHOS and remained stuck in a glycolytic program, diminishing T-cell metabolic fitness. In this regard, our data are in line with previous reports on T-cell metabolism showing a correlation between metabolic plasticity and acquisition of activated phenotype.1,2 Second, pseudohypoxic T cells upregulated the enzymatic machinery to produce (and sense) adenosine in an autocrine manner. Numerous reports have highlighted the ability of cancer cells to release adenosine during hypoxia or even in normoxic environments because of genetic mutations or production of oncometabolites by tumor cells.14 Here, we show that T cells upregulated the machinery for adenosine production themselves to detoxify the extracellular ATP, which is a hallmark of hypoxia and glycolytic metabolism.23 During glycolysis, ATP is rapidly produced and leaks out from the cell,23 constituting the substrate for the production of adenosine.13 This enzymatic conversion determines a shift from a proinflammatory ATP-driven milieu to an anti-inflammatory TME.38 Production of extracellular adenosine negatively affected T-cell function by dampening TCR signaling. The purinergic signaling, which is evolutionarily well conserved, has been proposed as a metabolic checkpoint aimed at preventing excessive inflammation.39-41 In light of this, pseudohypoxia generated a nonresolved inflammatory setting, characterized by forced glycolysis, which usually characterizes inflammatory T-cell subsets,42 and adenosine production to counteract this inflammation,41 both reducing antitumor immunity.
The role of hypoxia as a means by which CLL cells acquire a survival advantage in the lymph node and simultaneously generate a tumor-permissive immune milieu was investigated by Deaglio et al, who applied hypoxia to mimic the lymph node environment.17 As anticipated previously, hypoxia is a hallmark of secondary lymphoid organs, whereas blood is oxygenated.18,43 Here, we show, in a general sense, that T-cell hypoxic signaling not only derives from scarcity of oxygen in the TME, but it is also intrinsically determined by the dialogue with leukemic cells inducing metabolic alterations. Intriguingly, it was reported that signals in the lymph node, such as activated B-cell receptor signaling in the CLL cells, results in increased fraction of CD4+CD39+ T cells, as exemplified by B-cell receptor stimulation of PBMCs from patients with CLL.43 The authors speculate this might derive from hypoxia, correlating with tumor burden and, therefore, with prognostic significance.
Although a hypoxic-like gene signature as well as increased expression of CD39 was found in T cells from patients with CLL in the presence of the autologous CLL counterpart, we did not observe increased levels of HIF-1α. Besides being a well-established HIF-1α target,44 CD39 is also regulated by the transcription factor SP1,39 which is also strongly linked to hypoxic gene transcription.45 Alternatively, HIF-1α could be activated in vivo during tumorigenesis, priming T cells for dysfunction, a process that could be dynamically recapitulated in the coculture model.
Our data suggest that PGA-1 and CLL-derived cell lines induce Treg cell formation. In support, increased numbers of Treg cells in patients with CLL have been reported and were considered a marker of clinical significance.46 Human FOXP3+ Treg cells coexpress CD39 and CD73,31,38,47 which are increasingly designated as markers for this subset. It is also believed that adenosine conveys a substantial part of the immunosuppressive function of Treg cells.30 In a recent study, hypoxic features and CD39 marked a terminal exhausted phenotype, which interestingly shared similar immunosuppressive features of Treg cells.48
Our findings are in line with previous reports indicating the unfavorable effects of systemic hypoxia on antitumor immunity,10,17,49 in contrast to acute induction of hypoxia in vitro, which is beneficial in T-cell expansion studies. HIF-1 itself is considered both positive and negative regulator of T-cell activity,37,50,51,52 and it is currently unknown whether its role is defined by time-dependent activation or by its different ways of regulation, including (pseudo)hypoxia preventing protein degradation vs TCR-induced transcriptional upregulation and mTOR-mediated increase in protein synthesis.24
Several aspects of T-cell dysfunction by pseudohypoxia could be amenable to therapeutic targeting. In light of the role of HIF-1α in T-cell activation summarized earlier, it might be counterproductive to use HIF-1α inhibitors as a therapeutic approach. Instead, a viable option would be to target hypoxia-driven mechanisms that ultimately result in immunosuppression, such as the skewing of pyruvate to lactate, adenosine production, and/or the purinergic pathway. For example, interfering with purinergic signaling by using Etrumadent/AB928 was able to restore T-cell activation in our study. This A2A/BR antagonist is currently under investigation in early phase clinical trials for the treatment of solid tumors but has not been tested yet in hematological malignancies. Our data suggest a possible window for this molecule or other A2A/BR antagonists in the treatment of CLL. It was recently reported that AB928 showed beneficial effect on chimeric antigen receptor T-cell functioning,53 offering promising results to ameliorate T-cell dysfunction to improve this promising treatment modality. Moreover, small molecule inhibitors as well as monoclonal antibodies targeting CD39 and CD73 are available and could potentially promote T-cell immunity against tumor cells.54 Another therapeutic strategy might be to target T-cell metabolic alterations. For example, rerouting pyruvate to OXPHOS by using antimetabolites could improve T-cell activation. It has been described that hypoxia promotes the assembly of the multienzymatic complex purinosome,55 which carries out the de novo synthesis of purines. Other purines besides adenosine, such as inosine or guanosine, were described to activate A2A/BR in various reports.41,56,57 In the context of T-cell dysfunction, these purines synthetized by the purinosome upon hypoxia, when found in extracellular environment, may also negatively affect T-cell function.
With regard to the limitations of our study, we are aware that our model system and the cell lines used might not reflect important biologic features of CLL biology. Moreover, concerns regarding mixed lymphocyte reaction might arise, but we did not observe any significant proliferation in our short-term culture assays. Our coculture model is not susceptible to patient variability, but confirming the findings of our model with primary PBMCs from patients with CLL was an essential component of our study.
In conclusion, leukemic cells of CLL origin enforce a pseudohypoxic state on T cells with negative outcomes for T-cell immunity. Our data demonstrate the positive impact of targeting the purinergic axis to improve T-cell function in a (pseudo)hypoxic environment. This novel mechanistic insight suggests that modulation of hypoxia and purinergic pathways might be an efficient strategy to overcome T-cell dysfunction in CLL and possibly other malignancies and improve the outcome of T-cell based immunotherapy.
Acknowledgments
The authors thank the HDs and patients for their blood donations and cooperation in the studies and Faris Naji from Tercen for the discussion about metabolomics data. This work was supported by the EU’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie Grant Agreement 766214 (META-CAN); ERC Consolidator BOOTCAMP (864815); Lymph and Co 2018-LYCo-008; and Cancer Center Amsterdam grant 2022.
Authorship
Contribution: C.M., H.S.-M., and E.E. designed the research; C.M., G.C., C.J., H.S.-M., B.V.S., and M.v.W. performed research; C.M., C.J., G.C., H.S.-M., and F.S.P. analyzed data; A.P.K. provided patient samples and reviewed the manuscript; and C.M., C.J., and E.E. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric Eldering, Department of Experimental Immunology, Amsterdam UMC, Location AMC, 1105 AZ Amsterdam, The Netherlands; e-mail: e.eldering@amsterdamumc.nl.
References
Author notes
Transcriptomic data are deposited in the Gene Expression Omnibus database (accession number GSE226599).
Data are available on request from the corresponding author, Eric Eldering (e.eldering@amsterdamumc.nl).
The full-text version of this article contains a data supplement.

![T-cell function is dampened in parallel to deregulated metabolism in the presence of malignant B cells, especially from CLL origin. PBMCs from HDs were thawed and cultured in the presence or absence of PGA-1, CII, and Mec-1 derived from CLL; LME-1, U-266, and RPMI-8226 derived from MM; and Nalm-6 cells derived from acute B-lymphoblastic leukemia. After 24 hours, anti-CD3/CD28 antibodies were added, and T cells were analyzed 48 hours after stimulation. (A) PBMCs were stained with carboxyfluorescein succinimidyl ester, and CD4+ T-cell proliferation was assessed. (B) Brefeldin A and Golgi Stop were added to the coculture 4 hours before analysis to assess cytokine production and degranulation. Percentage of CD4+ T cells producing tumor necrosis factor (TNF)-α, interleukin-2 (IL-2), and interferon-gamma (IFNγ) 48 hours after stimulation and percentage of cells expressing CD107A at the cell surface is plotted. After gating on CD4+ T cells, activated T cells (CD4+CD25+) were analyzed for (C) T-cell activation, represented by CD25 levels (gMFI); metabolic capacity, as indicated by (D) glucose uptake (2-NBDG); and (E) mitochondrial mass (MitoTracker green FM). (F) Activated T cells were analyzed for the expression of CD25 and (G) effector function after coculture with CII, Mec-1, U-266, and RPMI-8226. Data are presented as mean ± standard error of the mean (SEM). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05; ns, not significant (1-way analysis of variance [ANOVA] followed by Tukey multiple comparison test or paired t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/21/10.1182_bloodadvances.2023010305/3/m_blooda_adv-2023-010305-gr1.jpeg?Expires=1769249539&Signature=wd2ikEkWrucqaqfY55FDzJkkJ2~l4m4eHCeZujLgN7GJ2RbglpXyVJ1ivgbgR-vjroV-ODQz6i1GdIIztrs604M-guH-0tJ-AgP~Lr1xRnEtRA9vp7reOTHkvZOZT9yZseB8FbSuT4k2BB4TGtMtScI6HTYcGFjbEBUzC8kcmLlMYE5JgwJNjvgbKKNSV32scJIbFL32QsuXKN2pSKdgS-HruoM7jepy1dzYtRTJCNVynTWYdoNhyATcUadewdjEzs2brH2NWnVkDF26CHViqNaBa8BH8ZwnXZhHWuZhG-yifOKQi139qTwurEk9NCuPZyHKsDirAzSJK3w8GH~PJg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)