Key Points
KDM6A inactivation sensitizes T-ALL cells to glucocorticoids, enforces JDP2 expression, reverses this phenotype, and causes resistance.
Alterations in KDM6A and JDP2 modulate glucocorticoid-induced NR3C1 mRNA and glucocorticoid receptor protein expression in T-ALL cells.
Abstract
Glucocorticoids (GCs) are the cornerstone of acute lymphoblastic leukemia (ALL) therapy. Although mutations in NR3C1, which encodes the GC receptor (GR), and other genes involved in GC signaling occur at relapse, additional mechanisms of adaptive GC resistance are uncertain. We transplanted and treated 10 primary mouse T-lineage acute lymphoblastic leukemias (T-ALLs) initiated by retroviral insertional mutagenesis with GC dexamethasone (DEX). Multiple distinct relapsed clones from 1 such leukemia (T-ALL 8633) exhibited discrete retroviral integrations that upregulated Jdp2 expression. This leukemia harbored a Kdm6a mutation. In the human T-ALL cell line CCRF-CEM, enforced JDP2 overexpression conferred GC resistance, whereas KDM6A inactivation unexpectedly enhanced GC sensitivity. In the context of KDM6A knockout, JDP2 overexpression induced profound GC resistance, counteracting the sensitization conferred by KDM6A loss. These resistant “double mutant” cells with combined KDM6A loss and JDP2 overexpression exhibited decreased NR3C1 mRNA and GR protein upregulation upon DEX exposure. Analysis of paired samples from 2 patients with KDM6A-mutant T-ALL in a relapsed pediatric ALL cohort revealed a somatic NR3C1 mutation at relapse in 1 patient and a markedly elevated JDP2 expression in the other. Together, these data implicate JDP2 overexpression as a mechanism of adaptive GC resistance in T-ALL, which functionally interacts with KDM6A inactivation.
Introduction
A poor initial response to glucocorticoids (GCs) is a strong predictor of relapse in T-lineage acute lymphoblastic leukemia (T-ALL).1 Acquired mutations in GC pathway genes (eg, NR3C1 and NR3C2) are a well-characterized GC resistance mechanism in ALL2,3 and other causes such as NSD24 and CELSR25 mutations have recently emerged. In an unbiased screen to uncover GC resistance mechanisms, we transplanted 10 primary mouse T-ALLs generated by retroviral insertional mutagenesis into recipients and treated them with GC dexamethasone (DEX) with or without the pan-PI3 kinase inhibitor GDC-0941 until relapse.2 GR protein expression was reduced in ∼40% of relapsed leukemias and was associated with preexisting or acquired mutations in Nr3c1. However, the mechanisms underlying DEX resistance have not been characterized in the remaining ∼60% of relapsed T-ALLs.2
One of these leukemias (T-ALL 8633) exhibited different retroviral integrations near the Jdp2 locus in multiple recipients that relapsed during continuous treatment with DEX.2Jdp2, which encodes an epigenetic modifier and represses AP-1-mediated transcription, has been implicated as a T-ALL oncogene that induces intrinsic GC resistance in zebrafish thymocytes.6 T-ALL 8633 was the only leukemia in our panel with a Kdm6a mutation, a tumor suppressor gene that is mutated in hematologic and solid cancers, including ∼5% of pediatric and adult T-ALLs.7,8KDM6A encodes a demethylase for di- and tri-methylated histone 3 lysine 27 (H3K27me2/3), which physically associates with KMT2C (MLL3) and KMT2D (MLL4) methyltransferases. Human and mouse KDM6A share 97% sequence identity and the proline-to-serine mutation at codon 828 found in T-ALL 8633 is predicted to be deleterious7 (rank scores 0.77 and 0.72, respectively, using the latest 2 ensemble algorithms, REVEL and BayesDel). Based on these observations, we investigated the effects of KDM6A inactivation and increased JDP2 expression in a human T-ALL cell line and tested the hypothesis that these mutations might interact to modulate the sensitivity of T-ALL cells to GCs.
Methods
T-ALLs that were generated by injecting newborn mice with the MOL4070 retrovirus were transplanted and treated as described previously.2,9,10 Published methods were used to perform Southern blotting, CRISPR/Cas9 gene editing, DEX dose-response assays, sequencing, and western blotting.2 CCRF-CEM cell subclones S1 and S19 were isolated using limiting dilution. A pCDH-MSCV-T2A-mCherry lentiviral vector was used to overexpress JDP2, and mCherry-positive cells were isolated using a Sony SH800S cell sorter. JDP2 overexpression was confirmed in mCherry-positive cells using quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blotting. RT-PCR was performed on a QuantStudio 5 Real-time PCR Instrument (Applied Biosystems). Ribonucleoprotein based CRISPR editing was used to inactivate KDM6A (sgRNA TCTTTGTATGAACAGCTGGG). Homozygous mutant single cells were isolated via limiting dilution, prioritized using the online Synthego ICE prediction tool, and confirmed using Sanger sequencing and western blotting. GraphPad Prism software was used to generate Kaplan-Meier curves. All experiments were independently performed in triplicate, yielding consistent data. Data from independent experiments were pooled and analyzed using 2-way analysis of covariance (ANCOVA), as described in detail in supplemental Materials.
Results and discussion
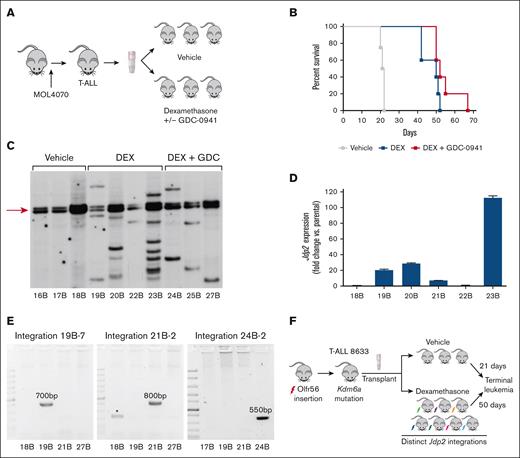
In this T-ALL model, retroviral insertions cooperate with somatic mutations in known human oncogenes and tumor suppressors such as Notch1 and Ikzf1.10,11 To identify the mechanisms of GC resistance, we transplanted 10 independent leukemias into recipient mice and treated them with vehicle, DEX, or DEX/GDC-0941 until relapse (Figure 1A).2 In T-ALL 8633, DEX treatment significantly prolonged survival, which was not enhanced by cotreatment with GDC-0941 (Figure 1B). Southern blot analysis of bone marrow extracted from 7 recipients that received DEX or DEX + GDC-0941 revealed MOL4070 integrations that were not detected in leukemias isolated from vehicle-treated mice (Figure 1C). Relapsed leukemias with this type of clonal evolution invariably exhibits intrinsic drug resistance.2,12-14 Shotgun cloning of inverse PCR products from multiple DEX-treated recipients of T-ALL 8633 identified viral integrations near the Jdp2 gene (supplemental Table 1), a known insertion site in retrovirally-induced ALL.15,16 These integrations were associated with elevated Jdp2 mRNA expression (Figure 1D). Consistent with the variable sizes of DNA fragments observed by Southern blot (Figure 1C), each Jdp2 integration was mapped to a distinct genomic locus. Given that MOL4070 is a replication-competent virus, we hypothesized that these integrations were acquired independently during DEX treatment.
DEX treatment extends the survival of recipients of T-ALL 8633 and results in the emergence of resistant subclones with diverse Jdp2 integrations. (A) Overview of T-ALL generation in mice infected with the MOL4070 retrovirus and subsequent transplantation and treatment of primary leukemia cells. (B) Kaplan-Meier survival analysis of T-ALL 8633 treated with vehicle (n = 4), DEX (n = 5) or DEX/GDC-0941 (n = 5) (vehicle vs DEX, P < .005; vehicle vs DEX/GDC-0941, P < .005; DEX vs DEX/GDC-0941, P > .05). (C) Southern blot showing patterns of MOL4070 integrations in genomic DNA extracted from leukemia cells harvested from individual moribund recipients of T-ALL 8633 at relapse, after treatment with vehicle, DEX, or DEX/GDC-0941. The arrow highlights the integration observed across all clones, which likely represents Olfr56 (see supplemental Table 1). (D) Jdp2 mRNA expression in parental/vehicle-treated (18B) leukemia 8633 and leukemias isolated from the 5 recipient mice treated with DEX at relapse (19B-23B). Error bars represent the standard deviation of technical triplicates. 22B is the only DEX-treated 8633 subclone without 1 or more Jdp2 integrations. (E) PCR amplification of unique host/virus DNA junction fragments detected by shotgun cloning in relapsed clones 19B, 21B, and 24B based on the data shown in supplemental Table 1. PCR products of the predicted sizes were only amplified from 19B (left), 21B (middle), and 24B (right) using primer pairs specific for each respective integration, which were sequence verified, as shown in supplementary Figs. 1 and 2. The asterisk in the middle panel indicates a background amplification product in 18B, which does not contain the junction fragment observed in 21B. (F) Schematic of the clonal evolution observed in murine RIM-induced leukemogenesis and preclinical trials. Molecular analysis of T-ALL 8633 revealed the same Olfr56 retoviral integration (red lightening bolt, arrow in Figure 1C and supplemental Table 1) and somatic Kdm6a mutation in all vehicle- and DEX-treated leukemias. After treatment with DEX, which significantly prolonged survival (Figure 1B), relapsed leukemia exhibited clonal evolution on Southern blots (Figure 1C) with distinct Jdp2 integrations (colored lightening bolts) that were likely acquired during in vivo treatment (Figure 1E).
DEX treatment extends the survival of recipients of T-ALL 8633 and results in the emergence of resistant subclones with diverse Jdp2 integrations. (A) Overview of T-ALL generation in mice infected with the MOL4070 retrovirus and subsequent transplantation and treatment of primary leukemia cells. (B) Kaplan-Meier survival analysis of T-ALL 8633 treated with vehicle (n = 4), DEX (n = 5) or DEX/GDC-0941 (n = 5) (vehicle vs DEX, P < .005; vehicle vs DEX/GDC-0941, P < .005; DEX vs DEX/GDC-0941, P > .05). (C) Southern blot showing patterns of MOL4070 integrations in genomic DNA extracted from leukemia cells harvested from individual moribund recipients of T-ALL 8633 at relapse, after treatment with vehicle, DEX, or DEX/GDC-0941. The arrow highlights the integration observed across all clones, which likely represents Olfr56 (see supplemental Table 1). (D) Jdp2 mRNA expression in parental/vehicle-treated (18B) leukemia 8633 and leukemias isolated from the 5 recipient mice treated with DEX at relapse (19B-23B). Error bars represent the standard deviation of technical triplicates. 22B is the only DEX-treated 8633 subclone without 1 or more Jdp2 integrations. (E) PCR amplification of unique host/virus DNA junction fragments detected by shotgun cloning in relapsed clones 19B, 21B, and 24B based on the data shown in supplemental Table 1. PCR products of the predicted sizes were only amplified from 19B (left), 21B (middle), and 24B (right) using primer pairs specific for each respective integration, which were sequence verified, as shown in supplementary Figs. 1 and 2. The asterisk in the middle panel indicates a background amplification product in 18B, which does not contain the junction fragment observed in 21B. (F) Schematic of the clonal evolution observed in murine RIM-induced leukemogenesis and preclinical trials. Molecular analysis of T-ALL 8633 revealed the same Olfr56 retoviral integration (red lightening bolt, arrow in Figure 1C and supplemental Table 1) and somatic Kdm6a mutation in all vehicle- and DEX-treated leukemias. After treatment with DEX, which significantly prolonged survival (Figure 1B), relapsed leukemia exhibited clonal evolution on Southern blots (Figure 1C) with distinct Jdp2 integrations (colored lightening bolts) that were likely acquired during in vivo treatment (Figure 1E).
To address this question, we selected unique Jdp2 integration sites in relapsed clones 19B, 21B, and 24B, designed primer pairs spanning the respective virus-host DNA junctions, and performed PCR amplification (supplemental Figure 1). These studies showed that each integration site was only detected in the relapsed sample from which it was originally isolated and was absent in DNA isolated from other relapsed samples and vehicle-treated recipients of T-ALL 8633 (Figure 1E; supplemental Figure 2). In contrast, targeted sequencing of 6 relapsed leukemias showed that all harbored the same Kdm6a mutation originally identified in T-ALL 8633 before treatment (data not shown). Altogether, these data are consistent with a model whereby a “founder” Olfr56 retroviral integration and the somatic Kdm6a mutation cooperated in leukemogenesis, with subsequent DEX treatment selecting for the outgrowth of rare leukemia cells with de novo Jdp2 integrations at relapse (Figure 1F).
To explore the possible functional interaction between Jdp2 and Kdm6a in modulating the GC response and resistance, we performed lentiviral transduction to overexpress JDP2, followed by CRISPR/Cas-9 gene editing to inactivate KDM6A in CCRF-CEM T-ALL cells. The human T-ALL cell line CCRF-CEM is sensitive to GCs and has low basal JDP2 expression.6 Two independent CCRF-CEM cell subclones (S1 and S19) were isolated by limiting dilution and transduced with either an empty pCDH-MSCV-T2A-mCherry vector (EV) or the same lentiviral vector encoding JDP2. These cells were referred to as EV (EV controls) and JDP (JDP2-expressing). CRISPR/Cas9 gene editing was used to disrupt KDM6A in both genotypes of each subclone (S1-EV, S19-EV, S1-JDP, and S19-JDP), and single-cell clones were isolated by limiting dilution to produce independent replicates for each genotype. These KDM6A knockout clones are referred to as EV/KDM (KDM6A knockout only) or JDP/KDM (JDP2 overexpressing; KDM6A knockout). The JDP2 mRNA expression and JDP2 and KDM6A protein levels are shown in supplemental Figure 3.
JDP2 overexpression conferred DEX resistance in wild-type CCRF-CEM cells (Figure 2A, JDP vs EV; P < .0001), whereas KDM6A knockout augmented DEX sensitivity (in 3 independent EV/KDM clones) [EV/KDM vs EV; P < .0001; supplemental Table 2A]. In contrast, CCRF-CEM cells with both KDM6A inactivation and JDP2 overexpression (JDP/KDM) were most resistant to DEX treatment (Figure 2A; supplemental Table 2A). Thus, JDP2 overexpression fully reversed the enhanced sensitivity of the KDM6A-mutant CCRF-CEM cells to DEX. In fact, JDP2-associated resistance in the KDM6A-mutant background (JDP/KDM vs EV/KDM) was significantly greater than JDP2-associated resistance in the KDM6A-WT setting (Figure 2A, JDP vs EV; cumulative difference in sensitivity to DEX across dose range −55.7 vs −212.2; P < .0001).
DEX sensitivity and GC pathway activation in CCRF-CEM cells and JDP2 expression in paired diagnostic/relapsed human pediatric T-ALLs. (A) Cell viability assay was assessed by Hoechst live/dead stain using flow cytometry. Individual data points for all the S1/S19 clones are shown. JDP2 overexpression caused moderate DEX resistance (JDP [blue] vs EV [gray], P < .0001 by ANCOVA). KDM6A inactivation significantly increased DEX sensitivity (EV/KDM [pink] vs EV [gray]; P <.0001) and JDP2 overexpression reversed this phenotype and conferred profound resistance (JDP/KDM [green] vs EV/KDM [pink]; P <.0001). See supplemental Table 2 for additional statistical analyses. (B) NR3C1 mRNA expression in untreated CCRF-CEM S1 and S19 cells and in cells exposed to 1 μM of DEX for 18 hours. NR3C1 is upregulated in the presence of DEX in all genotypes (P < .0001 by ANCOVA). JDP2 overexpression in the background of KDM6A inactivation resulted in a marked reduction in responsiveness to DEX (JDP/KDM vs EV/KDM; P <.0001). See supplemental Table 2 for additional statistical analyses. (C) Western blot showing glucocorticoid receptor protein levels in cells exposed to either control vehicle or DEX (500 nM) for 24 hours in all CCRF-CEM S1 and S19 subclones. (D) Change in JDP2 expression from diagnosis to relapse in 9 paired human T-ALL samples, including 2 with a KDM6A mutation. As shown, T-ALL with a V1112 frameshift KDM6A mutation had a unique ∼10-fold increase in JDP2 expression, and leukemia with a V1113_S1114 frameshift KDM6A mutation acquired an NR3C1 mutation at relapse.
DEX sensitivity and GC pathway activation in CCRF-CEM cells and JDP2 expression in paired diagnostic/relapsed human pediatric T-ALLs. (A) Cell viability assay was assessed by Hoechst live/dead stain using flow cytometry. Individual data points for all the S1/S19 clones are shown. JDP2 overexpression caused moderate DEX resistance (JDP [blue] vs EV [gray], P < .0001 by ANCOVA). KDM6A inactivation significantly increased DEX sensitivity (EV/KDM [pink] vs EV [gray]; P <.0001) and JDP2 overexpression reversed this phenotype and conferred profound resistance (JDP/KDM [green] vs EV/KDM [pink]; P <.0001). See supplemental Table 2 for additional statistical analyses. (B) NR3C1 mRNA expression in untreated CCRF-CEM S1 and S19 cells and in cells exposed to 1 μM of DEX for 18 hours. NR3C1 is upregulated in the presence of DEX in all genotypes (P < .0001 by ANCOVA). JDP2 overexpression in the background of KDM6A inactivation resulted in a marked reduction in responsiveness to DEX (JDP/KDM vs EV/KDM; P <.0001). See supplemental Table 2 for additional statistical analyses. (C) Western blot showing glucocorticoid receptor protein levels in cells exposed to either control vehicle or DEX (500 nM) for 24 hours in all CCRF-CEM S1 and S19 subclones. (D) Change in JDP2 expression from diagnosis to relapse in 9 paired human T-ALL samples, including 2 with a KDM6A mutation. As shown, T-ALL with a V1112 frameshift KDM6A mutation had a unique ∼10-fold increase in JDP2 expression, and leukemia with a V1113_S1114 frameshift KDM6A mutation acquired an NR3C1 mutation at relapse.
Next, we assessed the NR3C1 mRNA and GR protein expression levels. As expected, DEX treatment induced significant NR3C1 upregulation in all genotypes, with a trend toward higher NR3C1 mRNA levels in DEX-treated EV/KDM clones and less upregulation in JDP cells (Figure 2B, supplemental Table 2B). Consistent with the effects of DEX treatment on cell growth, the induction of NR3C1 expression was significantly reduced in JDP/KDM vs EV/KDM CCRF-CEM clones (P < .0001; Figure 2B; supplemental Table 2B). The GR protein expression followed a similar trend (Figure 2C). Interestingly, 1 of the 4 independent JDP/KDM clones (S1-JDP/KDM[2]) was highly resistant to DEX (Figure 2A, upper cluster of green dots) and failed to induce GR protein expression upon DEX exposure (Figure 2C). MCL-1 protein levels were similar in cells of all genotypes (supplemental Figure 4A-B). However, the induction of the proapoptotic factor BIM followed a pattern similar to that observed for changes in NR3C1 mRNA levels (supplemental Figure 4C), although these changes were not statistically significant (supplemental Table 2C).
Our studies on primary murine T-ALL 8633 and engineered CCRF-CEM cells suggest that DEX treatment might impose strong selective pressure on KDM6A-mutant T-ALLs to develop GC resistance. To address this question further, we analyzed data from a comprehensive serial genomic analysis of 103 pediatric patients with ALL that compared diagnostic and relapsed samples.3 Of the 9 patients with T-ALL and available transcriptome data from both diagnosis and relapse, 2 males harbored loss-of-function KDM6A mutations. Interestingly, the relapsed leukemias in both patients showed molecular evolution linked to GC resistance. Specifically, 1 patient acquired a somatic NR3C1 mutation and the other uniquely showed ∼10-fold increase in JDP2 mRNA expression (Figure 2D). The transcriptome data from the latter case confirmed the contributions of both JDP2 alleles, suggesting that overexpression may be due to an epigenetic mechanism rather than a structural variant. This relapsed T-ALL also had reduced NR3C1 expression compared with the paired diagnostic sample (supplemental Table 3), which is consistent with our biochemical findings.
Mansour et al6 reported that JDP2 causes intrinsic GC resistance in zebrafish thymocytes, and our studies of relapsed mouse and human leukemias implicated elevated JDP2 expression as a cause of adaptive resistance during GC treatment. We further implicated KDM6A and JDP2 as co-modulators of GC sensitivity in T-ALL and identified a functional interaction between them, which was supported by patient data. Investigating a hypothesis generated in a primary murine T-ALL model, we unexpectedly found that KDM6A inactivation enhanced GC sensitivity, which could be overcome by JDP2 overexpression in CCRF-CEM cells. Taken together with the clonal evolution observed in preclinical trials, we hypothesized that GC treatment results in selective pressure for the outgrowth of GC-resistant clones (eg, via JDP2 overexpression or other mechanisms). We further implicated modifications in the GC-induced expression of NR3C1 and GR as mediators of GC sensitivity in T-ALL cells with alterations in KDM6A and/or JDP2. Other genetic and epigenetic mechanisms similarly converge on GR signaling in ALL relapse.2-5,17 The FDA-approved BCL-2 inhibitor venetoclax and H3K27me3 inhibition18 are rational therapeutic approaches for overcoming adaptive GC resistance in patients with relapsed KDM6A-mutant T-ALL.
Acknowledgments
This work was supported by awards from St. Baldrick's Foundation, Ty Louis Campbell Foundation, Chan Zuckerberg Biohub, and NIH Research Training in Childhood Cancer (5T32CA128583 [A.L.L]); from the Genentech Foundation and an NIH training grant T32GM14132 (L.K.M.); and a Postdoctoral Fellowship (PF-14-070-01-TBG) from the American Cancer Society, including a supplement from the Hillcrest Committee, and an Alex's Lemonade Stand Foundation Young Investigator Grant (A.M.W.). Additional funding was provided by St. Baldrick's Foundation research grants R37 CA72614 and U54CA196519 (K.S.), NIH K08 CA256489 (B.J.H.), and R01 CA216391 (J.Z.).
Authorship
Contribution: A.L., K.S., and A.W. designed the experiments; A.L., A.W., K.T., B.H., and L.M. performed the experiments and/or analyzed the data; S.W.B. and J.Z. performed the computational analysis of the human data set and produced the associated figure panel and supplemental table; M.K. performed statistical analyses as summarized in supplemental Table 2; and all authors contributed to the writing of the manuscript and reviewed it before submission.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
The current affiliation for A.M.W. is the Department of Laboratory Medicine, Seattle Children's Hospital, Seattle, WA.
Correspondence: Kevin Shannon, Helen Diller Family Cancer Research Building, 1450 3rd St, Room 240, San Francisco, CA 94158; e-mail: Kevin.shannon@ucsf.edu.
References
Author notes
#Presented in abstract form at the 64th annual meeting of the American Society of Hematology (10-13 December 2022).
For data sharing, contact the corresponding author, Kevin Shannon (Kevin.shannon@ucsf.edu).
The full-text version of this article contains a data supplement.

![DEX sensitivity and GC pathway activation in CCRF-CEM cells and JDP2 expression in paired diagnostic/relapsed human pediatric T-ALLs. (A) Cell viability assay was assessed by Hoechst live/dead stain using flow cytometry. Individual data points for all the S1/S19 clones are shown. JDP2 overexpression caused moderate DEX resistance (JDP [blue] vs EV [gray], P < .0001 by ANCOVA). KDM6A inactivation significantly increased DEX sensitivity (EV/KDM [pink] vs EV [gray]; P <.0001) and JDP2 overexpression reversed this phenotype and conferred profound resistance (JDP/KDM [green] vs EV/KDM [pink]; P <.0001). See supplemental Table 2 for additional statistical analyses. (B) NR3C1 mRNA expression in untreated CCRF-CEM S1 and S19 cells and in cells exposed to 1 μM of DEX for 18 hours. NR3C1 is upregulated in the presence of DEX in all genotypes (P < .0001 by ANCOVA). JDP2 overexpression in the background of KDM6A inactivation resulted in a marked reduction in responsiveness to DEX (JDP/KDM vs EV/KDM; P <.0001). See supplemental Table 2 for additional statistical analyses. (C) Western blot showing glucocorticoid receptor protein levels in cells exposed to either control vehicle or DEX (500 nM) for 24 hours in all CCRF-CEM S1 and S19 subclones. (D) Change in JDP2 expression from diagnosis to relapse in 9 paired human T-ALL samples, including 2 with a KDM6A mutation. As shown, T-ALL with a V1112 frameshift KDM6A mutation had a unique ∼10-fold increase in JDP2 expression, and leukemia with a V1113_S1114 frameshift KDM6A mutation acquired an NR3C1 mutation at relapse.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/14/10.1182_bloodadvances.2021006881/2/m_blooda_adv-2021-006881-gr2.jpeg?Expires=1769230686&Signature=LQzLCIcDz0ldUqqjhjDgVPgZ4rTJtBRzAfApX3scpoG2ARau31ZSjCI-0NC4K3mg3G8vgkr9Hg~UAeSzJMRy1Bg2O4I6hT4fqmY6Ad1LkxFid7mDOuWwkUbGj6s3l-56Rlg0RuQXRIvzDwf4gnV7VgiHATLFcdDk38-~oIVtuJI4WlPPQAgvm9yI5nnfuC5XCgFfjPdqvw7iPwiC6EqATjp9wiZlaQiSA~LeCaa~Gmk~JjW-GtBAJhoA6oBR8hxFab4UJ7r-zQwwN2Qq-ZosBPJhONc~vDb1S1-gOwzFJF4S1aX8DC-776fFMHsWRFPh5a3qNSxrrjZTdv7UfKQm1g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)