Key Points
Immune transcriptome response following vaccination of patients with CLL is largely unimpaired but does not predict humoral immune response.
Heterologous ChAd-BNT vaccination elicits early immune response in patients with CLL treated with Bruton tyrosine kinase or Bcl-2 inhibitors.
Abstract
Patients with chronic lymphocytic leukemia (CLL) treated with B-cell pathway inhibitors and anti-CD20 antibodies exhibit low humoral response rates following SARS-CoV-2 vaccination. To investigate this observation, a prospective single-institution study was conducted comparing peripheral blood mononuclear cell transcriptional response with antibody and T-cell response rates following heterologous BNT162b2/ChAdOx1 vaccination of 15 patients with CLL/small lymphocytic lymphoma (SLL). Two-dose antibody response rate was 40%, increasing to 53% after booster. Patients on Bruton tyrosine kinase inhibitor (BTKi) and venetoclax ± anti-CD20 antibody within 12 months of vaccination responded inferiorly compared with those under BTKi alone. The 2-dose–T-cell response rate was 80%, which increased to 93% after the booster dose. Key transcriptional findings were that interferon–mediated signaling activation including activation of the JAK-STAT pathway generally occurred within days of vaccination, but was independent from the magnitude of the antibody response. Increasing counts of IGHV genes were associated with B-cell reconstitution and improved humoral response rate in the vaccinated patients. T-cell responses in patients with CLL appeared independent of treatment status, whereas higher humoral response rate was associated with BTKi treatment and B-cell reconstitution. Boosting was particularly effective when intrinsic immune status was improved by CLL treatment. Limitations included studying a relatively small cohort, with different treatments and vaccination schedules.
Introduction
Patients with chronic lymphocytic leukemia (CLL) are considered to be at a high-risk for severe COVID-19 infection, mainly owing to their complex underlying immunodeficiency and inadequate immune response to infections.1-3 They not only suffer from immune dysregulation by the disease itself, but their immune system is further disrupted by treatment-related effects.4-6 Patients who are heavily pretreated with chemoimmunotherapy and exposed to anti-CD20 antibody or treated actively with B-cell pathway inhibitors experience suboptimal antibody response to COVID-19 vaccination compared with CLL treatment-naïve.7-13 Robust data on immunogenicity of 2-dose homologous or heterologous BNT162b2/ChAdOx1 vaccine schedules in patients with leukemia have demonstrated an enhanced humoral and/or cellular immune response.14,15 Heterologous vaccine schedules also enhance humoral response in individuals without hematological disease.16,17 The European Medicines Agency and the European Centre for Disease Prevention and Control discussed potential benefits of heterologous regimens in 2021.18
Although patients with CLL who received their last treatment within 12 months preceding standard vaccination program demonstrate low response rates, vaccine response rates increase in seronegative, actively treated patients following boosting.14,19 In addition, potential protection against COVID-19 infection provided by T cells, even in the absence of a humoral response, is of particular clinical interest.20,21 T-cell activation with release of interferon gamma (IFN-γ) following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is associated with mild disease and viral clearance.21,22 T-cell–mediated immune responses are reported in patients with lymphoid malignancies in the absence of a humoral response.23 However, in a mixed group of patients with cancer, they were documented more commonly in combination with a humoral response.24
Early responses to vaccination are elevated levels of interferons and other cytokines, which activate the JAK/STAT signaling pathway and induce expression of immediate and innate response genes. We have used RNA-seq of peripheral immune cells to identify the innate immune response of healthy individuals receiving the standard homologous BNT162b225 or a heterologous ChAdOx1/BNT162b217 regimen. Specific genetic pathways are differentially activated within the first 2 days after vaccination and more prominently in the heterologous cohort. However, there are no reports available in literature on the immune transcriptomic response in patients with CLL receiving COVID-19 vaccines.
The utility of heterologous vaccination regimens for improving immune response in immunocompromised patients continues to be deliberated.26,27 To add to this important discussion, here we provide a comprehensive transcriptome analysis of peripheral immune cells from patients with CLL who received heterologous ChAdOx1/BNT162b2 vaccination and monitored their innate and humoral immune response for 4 months following the third vaccination in combination with detailed discussion on disease status, treatment regimens, and response to COVID-19 infection during follow-up.
Methods
Ethical approval
Ethical approval (#20-225) to conduct this analysis was granted by the institutional review board of the Ludwig-Maximilian University, Munich as the responsible ethics committee. Written informed consent was obtained from the study participants.
Study population, study design, and recruitment
From June 2021 through July 2021, 15 patients diagnosed with CLL/SLL between 2003 and 2021 were recruited in a single institution (Department of Hematology and Infectious Diseases, Munich Clinic, Munich Schwabing, Germany). They received vaccination for SARS-CoV-2 per recommendation of booster vaccination by the Standing Committee on Vaccination (STIKO) at the Robert Koch Institute in Germany (Epidemiological Bulletin 39/2021).28 After providing written informed consent for data collection, 5 seronegative patients received a third (3-dose) after standard 2-dose homologous vaccination of BNT162b2 or ChAdOx1. All were heavily pretreated (Patients 103 and 104) or recently treated with anti-CD20 mAbs (Patients 105 and 106) or a Bruton tyrosine kinase inhibitor (BTKi) (Patient 107). In addition, 10 patients (2-dose) with different CLL disease and treatment status (Patients 201-213), half of whom were seropositive after prime dose of ChAdOx1, received a second homologous or heterologous dose. At the time of vaccination 14 of 15 patients did not have a history of COVID-19 infection. Between October 2021 and December 2021, all 10 patients of 2-dose received a BNT162b2 3-dose. Antibody response and incidence and outcome of COVID-19 infections were recorded per routine CLL management. Four patients (Patients 105, 107, 206, and 209) had a breakthrough COVID-19 infection with Omicron variant, documented by polymerase chain reaction testing, around 6 months after the third vaccination, all with mild symptoms. Patient 209 had a previous presumptive COVID-19 infection in January 2020 characterized by severe pneumonia with positive detection of anti-COVID-19 nucleocapsid antibodies. Patient 105 received antiviral treatment with molnupiravir.
QuantiFERON SARS-CoV-2 assay
SARS-CoV-2-specific T cells were analyzed using the QuantiFERON SARS-CoV-2 Research Use Only platform.29 The QuantiFERON SARS-CoV-2 Starter Pack (catalog number 626115; Qiagen), Extended Pack (catalog number 626215; Qiagen), and Control Set (catalog number 626015; Qiagen) were employed, consisting of assay tubes coated with 1 of the 3 sets of selected SARS-CoV-2 T-cell antigens: Ag1 - CD4+ T-cell epitopes from the S1 subunit (receptor binding domain) of the SARS-CoV-2 spike protein, Ag2 - CD4+ and CD8+ epitopes from the S1 and S2 subunits of the SARS-CoV-2 spike protein, and Ag3 (Extended Pack) - CD4+ and CD8+ epitopes from S1 and S2, as in Ag2, but also immunodominant CD8+ epitopes of the whole proteome. The Control pack contains a ‘Nil tube’ which serves as the negative control and a ‘Mitogen tube’ which serves as a positive control. Heparinized blood samples were transported to the laboratory within 4 hours of collection. A 1-mL sample was then transferred to each of the 3 SARS-CoV-2 blood collection tubes (SARS-CoV-2 specific antigens Ag1, Ag2, Ag3). After 24 hours of stimulation, plasma from the stimulated samples was used for the detection of IFN-γ. Detection was carried out using the QuantiFERON ELISA Human IFN-γ (Qiagen) kit. Detected IFN-γ cutoff level >0.1 IU/mL was evaluated as positive response.
SARS-CoV-2 antibody ELISA
End point binding immunoglobulin G (IgG) levels to the S1 domain of the spike protein of SARS-CoV-2 were measured using the semiquantitative Anti-SARS-CoV-2 ELISA IgG (Euroimmune, Lübeck, Germany), according to the manufacturer’s instructions. Positive responses included both IgG ratio ≥1.1 and borderline values of IgG between 0.8 and 1.0. Negative responses were IgG ratio <0.8. Cutoff values for lack of seroconversion (standard of active level) was set as <0.8. In addition, quantitative Anti-SARS-CoV-2 ELISA IgG measurement (Atellica IM SARS-CoV-2 IgG, Siemens) was performed with positive response ≥21.8 binding antibody unit/mL (BAU/mL) and negative <21.8 BAU/mL.
Virus neutralization test
SARS-CoV-2 (strain MUC IMB-1, clade B1) neutralizing antibody titers were determined as previously described,30 including positive and negative controls. Heat-inactivated serum samples in duplicates, including positive and negative control samples, were serially diluted in 96-well tissue culture plates starting at 1:5 to a maximum of 1:640. Virus stocks (50 TCID/50 μL) were prepared and stored at −80°C until further use. Virus was preincubated (1 hour at 37°C) with diluted serum samples before Vero E6 cells (1 × 104 cells/50 μL) were added. After 72 hours (37°C), supernatants were discarded and wells were fixed (13% formalin/ phosphate buffered saline [PBS]) and stained (crystal violet, 0.1%). The neutralizing antibody titer corresponded to the reciprocal of the highest serum dilution showing complete inhibition of cytopathic effect.
Flow cytometry
Cells were stained with fluorochrome-labeled antibodies according to the manufacturer’s instructions (Staining 1: CD19 PE-Cy7 and CD5 APC; Staining 2: CD3 FITC, CD4 APC-Cy7, CD8 Amcyan, PD1 PE-Cy7, CD25 PE, CD62L APC; staining 3: CD11c PE, CD14 APC-Cy7, HLA-DR PE-Cy7, CD56 PerCP710, all Biolegend). To block free Fc receptors human Fc receptor binding inhibitor polyclonal antibody (eBioscience) were added 10 minutes before labeled antibodies. Dead cells were excluded by DAPI (1 μg/mL) (Sigma Aldrich) staining. Flow cytometric analysis was performed using a FACS Canto II cytometer (BD Bioscience). Data were analyzed with the FlowJo software version 10.7.1 (BD Bioscience).
Extraction of the buffy coat and purification of RNA
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by density gradient centrifugation using Ficoll-Paque (GE Healthcare, Chicago, IL). CD19+ B-cells were depleted by magnetic-activated cell sorting using human CD19 MicroBeads (Miltenyi, Bergisch-Gladbach, Germany) if CLL cell population exceeded 10% of viable lymphocytes, as determined by flow cytometry, before RNA extraction. 3 × 106 PBMCs (with or without CD19 depletion) were collected, washed with PBS and resuspended in 200 uL Homo-TG buffer (Maxwell 16 LEV simplyRNA Purification Kit, Promega) and stored at –80°C. RNA was extracted on the Maxwell 16 Instrument according to manufacturer’s protocol and stored at –80°C for further processing.
mRNA sequencing (mRNA-seq) and data analysis
Bulk RNA-seq was performed on 3 million PBMCs obtained before the second vaccination (2-dose cohort) and third vaccination (3-dose cohort) and at days 1/2 (D1/2), 7 (D7), and week 4 to 5 (W4-5) after the vaccination. RNA-seq was conducted on a total of 42 samples with an average sequencing depth of at least 200 million reads per sample. The poly-A containing mRNA was purified by poly-T oligo hybridization from 1 μg of total RNA and cDNA was synthesized using random primers and SuperScript III (Invitrogen). Libraries for sequencing were prepared according to the manufacturer’s instructions with TruSeq Stranded mRNA Library Prep Kit (I RS-20020595; llumina) and paired-end sequencing was done with a NovaSeq 6000 instrument (Illumina) yielding 200 to 350 million reads per sample. The raw data were subjected to QC analyses using the FastQC tool (version 0.11.9) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). mRNA-seq read quality control was done using Trimmomatic31 (version 0.36) and STAR RNA-seq32 (version STAR 2.5.4a) using 150 bp paired-end mode was used to align the reads (hg19). HTSeq33 (version 0.9.1) was used to retrieve the raw counts and subsequently, Bioconductor package DESeq234 in R (https://www.R-project.org/) was employed to normalize the counts across samples and perform differential expression gene analysis. The RUVSeq35 package was applied to remove confounding factors. The data were prefiltered keeping only genes with at least 10 reads in total. The visualization was done using dplyr (https://CRAN.R-project.org/package=dplyr) and ggplot2.36 The genes with log2 fold change >1 or <−1 and adjusted P value (PAdj) <.05 corrected for multiple testing using the Benjamini-Hochberg method were considered significant and then a cgene-enrichment analysis was conducted (Gene Set Enrichment Analysis [GSEA], https://www.gsea-msigdb.org/gsea/msigdb). For T- or B-cell receptor repertoire sequencing analysis, trimmed fastq files from bulk RNA-seq were aligned against human V, D, and J gene sequences using the default settings with MiXCR.37,38 CDR3 sequence and the rearranged B-cell receptor (BCR)/T-cell receptor (TCR) genes were identified. Comparative reference cohorts were healthy individuals receiving heterologous (ChAd-BNT) or homologous (ChAd-ChAd) vaccinations.17
Statistical analysis
Differential expression gene (DEG) identification used Bioconductor package DESeq2 in R. P values were calculated using a paired, 2-side Wilcoxon test and adjusted PAdj corrected using the Benjamini–Hochberg method. Genes with log2 fold change >1 or <−1, PAdj <.05 and without 0 value from all sample were considered significant. For significance of each GSEA category, significantly regulated gene sets were evaluated with the Kolmogorov-Smirnov statistic. Fisher exact test was used to test for statistically significant associations between IgG seroconversion and positive IFN-γ release test in both the 2-dose or 3-dose vaccination groups and patient characteristics between responders and non-responders (version 9.4.1; GraphPad Prism, San Diego, CA). A value of ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001 was considered statistically significant.
Results
Patient characteristics
Patient baseline and characteristics are summarized in Table 1, Figure 1, and supplemental Table 1. At the time of the vaccination, 2 patients (13%) had treatment-naïve CLL. Seven (47%) were on treatment without remission (2 frontline, 4 relapse) or with remission (1 relapse). Six patients (40%) were off therapy, including 4 in clinical complete or partial remission (2 frontline, 2 relapse) and 2 on relapse, in need of treatment (1 frontline, 1 relapse). Of the treated patients, 4 received venetoclax monotherapy (Patients 103, 104, 201, and 203) and 3 ibrutinib monotherapy (Patients 107, 205, and 208). Six (40%) were off therapy, including 4 in clinical complete or partial remission (2 frontline, 2 relapse) and 2 on relapse, in need of treatment (1 frontline, 1 relapse). Eleven of 15 patients were previously on anti-CD20 monoclonal antibodies with/without chemotherapy, either more than 12 months (7 patients) or within 12 months (4 patients) before vaccination. Unfavorable prognostic CLL parameters included β2M >3.5 mg/L (2/15), complex karyotype (2/15), trisomy 12 (1/15), unmutated IGHV gene status (13/15), and presence of TP53/del(17p) and/or del(11q) (6/15). Median IgG level was 619 mg/dL (range, 159-1141), IgM level of 45 mg/dL (range, <5-179), and IgA level of 98 mg/dL (range, 12-210). Median absolute lymphocyte count was 5.9 per μL (range, 3.3-46.7). The median of B lymphocyte counts (%) was 7 (range, 0-84.6) (supplemental Table 2).
Characteristics of CLL study population
| N . | 15, n (%) . |
|---|---|
| Age (y), median (range) | 69 (59-82) |
| Gender | |
| Female | 6 (40) |
| Male | 9 (60) |
| Race | |
| Caucasian | 15 (100) |
| Disease status | |
| Naïve | 2 (13) |
| Frontline active | 2 (13) |
| Relapse active | 3 (20) |
| Relapse in need of treatment | 2 (13) |
| In remission | 5 (33) |
| Previous treatment | |
| <3 | 4 (27) |
| ≥3 | 7 (47) |
| Current treatment | |
| BTK inhibitor (BTKi) | 3 (20) |
| Bcl2 inhibitor (Bcl2i) | 4 (27) |
| Off-therapy | 6 (40) |
| Watch and wait (W&W) | 2 (13) |
| IGHV status | |
| Mutated | 2 (13) |
| Risk factors | |
| Trisomy 12 | 1 (7) |
| 17p deletion/T53 mutation | 5 (33) |
| 11q deletion | 2 (13) |
| Complex karyotype | 2 (13) |
| High βM2 | 2 (13) |
| Serum IgG level (mg/dL), median (range) | 619 (159-1141) |
| Serum IgM level (mg/dL), median (range) | 45 (<5-179) |
| Serum IgA level (mg/dL), median (range) | 98 (10-210) |
| B lymphocytes (%), median (range) | 7.2 (0-84.6) |
| N . | 15, n (%) . |
|---|---|
| Age (y), median (range) | 69 (59-82) |
| Gender | |
| Female | 6 (40) |
| Male | 9 (60) |
| Race | |
| Caucasian | 15 (100) |
| Disease status | |
| Naïve | 2 (13) |
| Frontline active | 2 (13) |
| Relapse active | 3 (20) |
| Relapse in need of treatment | 2 (13) |
| In remission | 5 (33) |
| Previous treatment | |
| <3 | 4 (27) |
| ≥3 | 7 (47) |
| Current treatment | |
| BTK inhibitor (BTKi) | 3 (20) |
| Bcl2 inhibitor (Bcl2i) | 4 (27) |
| Off-therapy | 6 (40) |
| Watch and wait (W&W) | 2 (13) |
| IGHV status | |
| Mutated | 2 (13) |
| Risk factors | |
| Trisomy 12 | 1 (7) |
| 17p deletion/T53 mutation | 5 (33) |
| 11q deletion | 2 (13) |
| Complex karyotype | 2 (13) |
| High βM2 | 2 (13) |
| Serum IgG level (mg/dL), median (range) | 619 (159-1141) |
| Serum IgM level (mg/dL), median (range) | 45 (<5-179) |
| Serum IgA level (mg/dL), median (range) | 98 (10-210) |
| B lymphocytes (%), median (range) | 7.2 (0-84.6) |
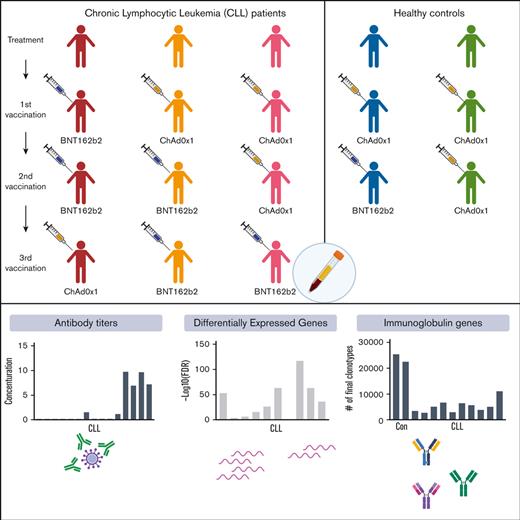
Study population and design. (A) CLL patient groups by treatment (naïve [n = 2], treatment active [n = 7], and off-treatment [n = 6]) and by drugs (BTKi [n = 3], Bcl2i [n = 4], anti-CD20 for less than 12 months [n = 4], and anti-CD20 for more than 12 months [n = 2]). (B) Blood samples were collected before the second (II-D0) and third (III-D0) vaccination and at days 2 (D2), 7 (D7) and 14 (D14) and weeks 4 to 5 (W4-5) after second and third vaccination as indicated by the colored circles. Mean and standard deviation for the number of days between first and second vaccinations was 66 ± 25 days and between second and third vaccinations was 131 ± 43 days. (C) The number of patients who received each vaccine regimen.
Study population and design. (A) CLL patient groups by treatment (naïve [n = 2], treatment active [n = 7], and off-treatment [n = 6]) and by drugs (BTKi [n = 3], Bcl2i [n = 4], anti-CD20 for less than 12 months [n = 4], and anti-CD20 for more than 12 months [n = 2]). (B) Blood samples were collected before the second (II-D0) and third (III-D0) vaccination and at days 2 (D2), 7 (D7) and 14 (D14) and weeks 4 to 5 (W4-5) after second and third vaccination as indicated by the colored circles. Mean and standard deviation for the number of days between first and second vaccinations was 66 ± 25 days and between second and third vaccinations was 131 ± 43 days. (C) The number of patients who received each vaccine regimen.
Antibody response
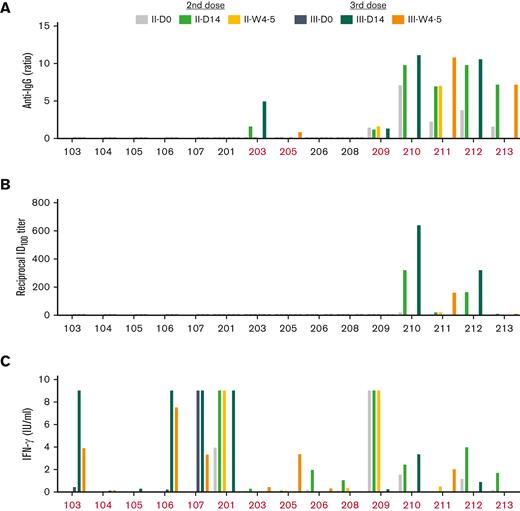
We analyzed antispike IgG antibody levels, neutralizing antibody levels and immune transcriptomes (RNA-seq) on peripheral immune cells before the vaccination, referred to as day (D) 0 (D0), on days 2 (D2), 7 (D7), and 14 (D14) and weeks 4 to 5 (W4-5) after vaccination (Figure 1B). Circulating antibody levels were most closely associated with treatment history.9,39 After 2-dose vaccination, antispike (S) IgG was detected in 6 of 15 patients (40%), including 2 of 6 (33%) who were off-therapy (Patients 210 and 212); 2 of 2 (100%) who were treatment-naïve (Patients 211 and 213); 1 of 4 (25%) who were on active venetoclax (203); none (0%) who were on ibrutinib; and 1 of 4 (25%) on anti-CD20 for less than 12 months (209) (Figures 2A and 3A; supplemental Table 3). Following 3 vaccination doses, 8 of 15 patients (53%) showed detectable anti-S antibodies (Figures 2A and 3A; supplemental Table 3). Several interesting responses occurred after a 3-dose heterologous regimen. Two naïve patients (Patients 211 and 213) that received frontline therapy with acalabrutinib shortly before or after their BNT162b2 3-dose retained seroconversion beyond 6 months after treatment. Patient 203, who received frontline treatment with venetoclax monotherapy before first ChAdOx1 vaccination, showed a detectable anti-S IgG response at D14 after second ChAdOx1 dose that was lost at W4-5 but then restored after a BNT162b2 3-dose. Patient 209, who had an undiagnosed COVID-19 infection before receiving first vaccination and stopped venetoclax+anti-CD20 treatment shortly before the second vaccination, showed an antibody response after first vaccination and retained detectable antibody levels through subsequential vaccinations, even with ongoing B-cell depletion in the context of CLL remission. Two patients on ibrutinib (Patients 107 and 205), who failed humoral response after 2 homologous ChAdOx1 doses, seroconverted after the third heterologous BNT162b2 dose. Patient 107, who was in second relapse and on ibrutinib treatment for 2 years, showed a delayed seroconversion 3 months after a temporal pause of ibrutinib while receiving a third heterologous BNT162b2 dose. Patient 205, who was on ibrutinib during the last 6 months, showed a delayed antibody response to the third heterologous BNT162b2 vaccination. Results were consistent with the notion that patients with adequate levels of serum immunoglobulins showed an increased antibody response after 3-dose vaccination, whereas those with very low serum levels of IgA, IgG, and IgM, respectively, failed to seroconvert following vaccination (supplemental Table 1). Overall, 7 of 15 of the patients failed to mount a detectable humoral response even after the 3-dose vaccination, irrespective of homologous or heterologous vaccination protocol. These patients were all heavily pretreated. Three were on current venetoclax treatment (103, 104, and 201), 1 presented with CLL relapse with therapy pending (Patient 106), 1 on prolonged ibrutinib (Patient 208), and 2 with anti-CD20 exposure less than 12 months before vaccination (Patients 105 and 206). Development of neutralizing antibodies was limited to the 4 patients showing a maximal antibody response following 2-dose vaccination that was not boosted by 3-dose vaccination (Patients 210, 211, 212, and 213) (Figure 2B; supplemental Table 3).
Humoral and T-cell immune responses following second dose and booster vaccination. (A) Plasma IgG antibody binding against the S1 domain of SARS-CoV-2 spike in patients with CLL. (B) Neutralizing antibody response to SARS-CoV-2 RBD (spike). (C) IFN-γ levels were measured from blood samples stimulated with a SARS-CoV-2 peptide cocktail containing antigens Ag1, Ag2, and Ag3 for 24 hours. IFN-γ level of >0.1 IU/mL was evaluated as positive response. Patients with a measurable response marked in red.
Humoral and T-cell immune responses following second dose and booster vaccination. (A) Plasma IgG antibody binding against the S1 domain of SARS-CoV-2 spike in patients with CLL. (B) Neutralizing antibody response to SARS-CoV-2 RBD (spike). (C) IFN-γ levels were measured from blood samples stimulated with a SARS-CoV-2 peptide cocktail containing antigens Ag1, Ag2, and Ag3 for 24 hours. IFN-γ level of >0.1 IU/mL was evaluated as positive response. Patients with a measurable response marked in red.
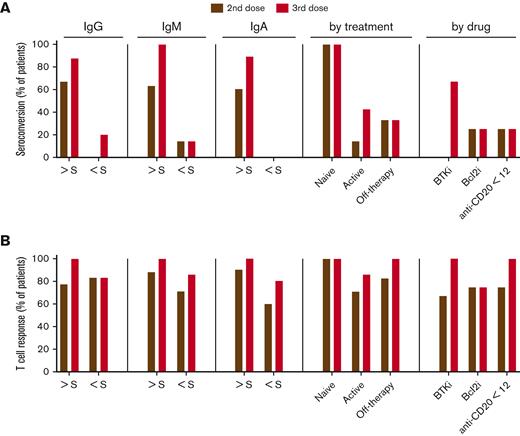
Antispike mediated T-cell response in patients with CLL. (A) Seroconversion rate. (B) T-cell response rate after 2-dose and 3-dose vaccines in patient subgroups based on clinical parameters and treatment status. Y-axis presents the proportions (%) of patients with CLL. S, standard of active level, IgG, 552 mg/dL; IgM, 33 mg/dL; and IgA, 69 mg/dL. Cutoff value for lack of antibody seroconversion (standard of active level) was set as <0.8. Cutoff value for a detectable IFN-γ response was >0.1 IU/mL.
Antispike mediated T-cell response in patients with CLL. (A) Seroconversion rate. (B) T-cell response rate after 2-dose and 3-dose vaccines in patient subgroups based on clinical parameters and treatment status. Y-axis presents the proportions (%) of patients with CLL. S, standard of active level, IgG, 552 mg/dL; IgM, 33 mg/dL; and IgA, 69 mg/dL. Cutoff value for lack of antibody seroconversion (standard of active level) was set as <0.8. Cutoff value for a detectable IFN-γ response was >0.1 IU/mL.
Cellular immune response
Robust T-cell responses were detected in 12 of 15 patients (80%) after 2-dose vaccination and 14 of 15 patients (90%) following 3-dose vaccination (Figure 2C; supplemental Table 4). T-cell response rate was independent of clinical characteristics, intrinsic immune and treatment status (Figure 3B). All seropositive patients (8/8) as well as 6 of 7 seronegative patients developed a T-cell mediated IFN-γ response. The 1 seronegative patient (Patient 104) without a T-cell response even after a 3-dose heterologous vaccination was on current venetoclax treatment. One patient (Patient 213), who started frontline therapy with acalabrutinib shortly before the 3-dose heterologous vaccination, lost T-cell response (supplemental Table 4). Further analysis of cellular immune-cell activation in all patients via flow cytometry revealed highly variable levels of T cells, myeloid-derived–suppressor cells (MDSCs), and NK cells was performed, but there was no correlation between these results and response rates detected by IFN-γ (supplemental Table 4).
Transcriptional immune response
Next, we assessed the vaccine-induced–innate immune response and transcriptional response in PBMCs isolated from 2- and 3-dose vaccination cohorts at days 0 (D0), 2 (D2), 7 (D7), and weeks 4 to 5 (W4-5) following vaccination (Figure 1B). The sequencing depth of 200 million reads per sample permitted an in-depth analysis of early response immunes and germline alleles induced by the vaccine. Reference cohorts were healthy individuals receiving heterologous (ChAd-BNT) or homologous (ChAd-ChAd) vaccinations.17 Samples were available for 8 patients: 3-dose BNT-BNT-ChAd (Patients 103, 104, and 106), 2-dose ChAd-ChAd (Patients 201, 208, and 209), 2-dose ChAd-BNT (Patients 206 and 212). First, numbers of DEGs were measured to examine the immediate response upon vaccination (Figure 4; supplemental Figures 1-2; supplemental Table 1). IFN-γ enrichment scores for DEGs at D2 were independent of antibody response. High levels were found both in patients with (Patients 209 and 212) and without (Patients 201 and 208) an antibody response (Figure 4A). Cell cycle pathway enrichment scores for DEGs were highest at D7, significantly higher both compared with D2 (Figure 4B) and D0 (Figure 4C). Similarly, higher enrichment in cell cycle pathways also did not correlate with seroconversion (Patients 106, 201, and 208). Patient 104, the 1 patient without a T-cell response, showed relatively low enrichment for both IFN-γ enrichment scores at D2 and cell cycle pathway enrichment at D7. Patient 206 had a higher IFN-γ score at D7 than at D2, suggesting delayed immune response. This patient exhibited a T-cell response and experienced a mild COVID-19 infection during follow-up, but this was not associated with seroconversion. Although the magnitude of activation varied, activation of interferon-induced genes (IFI27, ISG15, CXCL10, and GBP1), JAK/STAT signaling genes including STAT1, antiviral pattern recognition receptors (DDX58 and DHX58) and OAS family genes at D2 were qualitatively similar to healthy controls (Figure 4D). Induction of IFI6 and IFIT genes were higher in patients with CLL compared with healthy controls. Although the transcriptome response varied greatly between individual patients with CLL and was independent of the clinical characteristics and treatment status, the vast majority of patients with CLL regardless of antibody response exhibited an early transcriptome immune response presaging a later sustained T-cell mediated IFN-γ immune response.
Immune transcriptomes following vaccination. (A-C) Interferon-regulated genes are induced upon vaccination. Genes expressed at significantly higher levels at days 2 and 7 compared with those at day 0 were significantly enriched in Hallmark gene sets. X-axis denotes statistical significance as measured by minus logarithm of FDR q-values. Y-axis denotes ranked terms by q-values. (D) Heatmaps show log2 FC of IFN-γ response genes (top) and G2M target genes (bottom) significantly regulated between day 0 and day 2, day 2 and day 7, and day 0 and day 7. FDR, false discovery rate; G2M, G2/M checkpoint.
Immune transcriptomes following vaccination. (A-C) Interferon-regulated genes are induced upon vaccination. Genes expressed at significantly higher levels at days 2 and 7 compared with those at day 0 were significantly enriched in Hallmark gene sets. X-axis denotes statistical significance as measured by minus logarithm of FDR q-values. Y-axis denotes ranked terms by q-values. (D) Heatmaps show log2 FC of IFN-γ response genes (top) and G2M target genes (bottom) significantly regulated between day 0 and day 2, day 2 and day 7, and day 0 and day 7. FDR, false discovery rate; G2M, G2/M checkpoint.
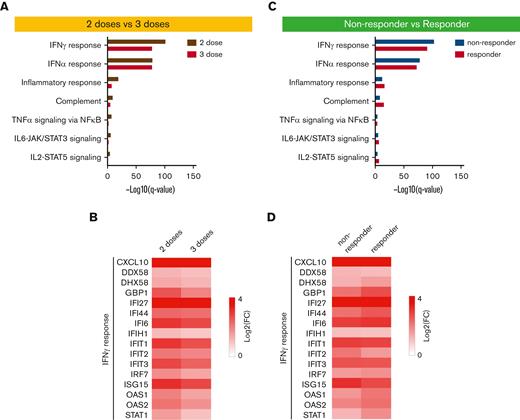
To further understand the differences by vaccine dosages and antibody response after vaccination, we compared 2-dose and 3-dose groups and non-responder vs responder groups (Figure 5; supplemental Tables 6-7). GSEA of significantly differential genes demonstrated that the genes activated within 2 days after second (Patients 210, 206,208, 209, and 212) or third (Patients 102, 104, and 106) doses were enriched in immune-response, interferon, and JAK-STAT pathways (Figure 5A). Key genes of the pathways were activated in both groups and were marginally higher in the 2-dose group (Figure 5B). The same enriched pathways were identified from the significantly activated genes within 2 days of vaccination in antibody non-responder (Patients 103, 104, 106, 201, 206, and 208) and responder (Patients 209 and 212) groups (Figure 5C). The induction folds of key genes were similar in both groups (Figure 5D).
Comparison of immune transcriptomes by vaccine doses as well as antibody response after vaccination. (A) Genes expressed at significantly higher levels between days of 2-dose and 3-dose groups were significantly enriched in Hallmark gene sets. X-axis denotes statistical significance as measured by minus logarithm of FDR q-values. Y-axis ranked the terms by q values. (B) Heatmaps showing log2 FC of significantly upregulated genes between day 0 and day 1 of 2-dose and 3-dose groups. (C) Hallmark gene sets of genes expressed at significantly higher levels between days of nonresponder and responder groups. (D) Heatmaps of genes significantly activated between day 0 and day 1 of 2-dose and 3-dose groups. FDR, false discovery rate.
Comparison of immune transcriptomes by vaccine doses as well as antibody response after vaccination. (A) Genes expressed at significantly higher levels between days of 2-dose and 3-dose groups were significantly enriched in Hallmark gene sets. X-axis denotes statistical significance as measured by minus logarithm of FDR q-values. Y-axis ranked the terms by q values. (B) Heatmaps showing log2 FC of significantly upregulated genes between day 0 and day 1 of 2-dose and 3-dose groups. (C) Hallmark gene sets of genes expressed at significantly higher levels between days of nonresponder and responder groups. (D) Heatmaps of genes significantly activated between day 0 and day 1 of 2-dose and 3-dose groups. FDR, false discovery rate.
Immunoglobulin germline repertoire
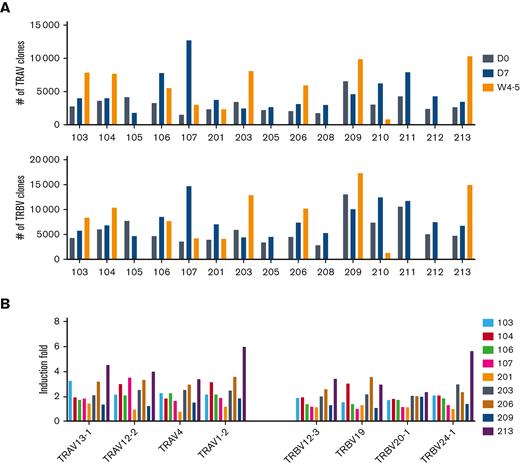
Finally, expression profiles of specific germline variable gene classes were interrogated. First, the range of immunoglobulin heavy-chain variable (IGHV), light chain (IGKV, IGLV), and T-cell receptor alpha/beta variable (TRAV, TRBV) gene usage in the patients with CLL was assessed (Figures 6-7). Final clonotype numbers were more than fourfold lower at D0 and D7 compared with that of vaccinated healthy controls (Figure 6A). A broad range of germlines in each patient was revealed by a deeper analysis of IGHV using complementarity determining regions (CDR)1 and CDR2 (Figure 6B). We observed IGHV3-74, IGHV3-30/IGHV3-33, IGHV1-18, IGHV3-23, IGHV3-21, and IGHV4-59 that are the basis of neutralizing antibodies identified in patients with SARS-CoV-2.40-43 In 6 patients, these clones were specifically increased (Patients 103, 104, 201, 203, 212, and 213). Three antibody responsive patients, including treatment-naïve (Patient 213), on venetoclax (Patient 203), and off-therapy (Patient 212), showed relatively higher numbers of IGHV clones. Because of transiently diminished B-cells upon active venetoclax treatment, Patient 203 had more IGHV clones at D0 and D7 than in W4-5 whereas other antibody responsive patients (Patients 209, 210, and 211) showed low and progressive numbers by recovered B cells. Notably, no BCR clones were detected in the antibody non-responder Patient 206, likely owing to completely depleted B cells by CLL treatment. Overall, results illustrate initially low levels of BCR in patients with CLL lacking a humoral and neutralizing antibody response gradually increase upon B-cell reconstitution with effective CLL therapy. In contrast to BCR genes, activation of TCR genes (TRAV, TRBV) was readily detected (Figure 7A), consistent with preservation of T-cell responses. Several TRAV and TRBV genes that are present in COVID-19 convalescent patients44,45 were induced in the patients with CLL at between D0 and W4-5 (Figure 7B).
SARS-CoV-2 B-cell memory. (A) Bar graphs show the number of clonotypes in each patient after the vaccination. (B) Pie charts for IGHV show the distribution of antibody sequences of individuals before the vaccination and after 7 days and 4 to 5 weeks. The number of sequences analyzed for individual are shown in the inner circle. Sizes of pie slices are proportional to the number of clonally related sequences. Persisting clones (same IGV genes) in both time points are shown as colored slices. Gray indicates sequences not overlapped between individuals.
SARS-CoV-2 B-cell memory. (A) Bar graphs show the number of clonotypes in each patient after the vaccination. (B) Pie charts for IGHV show the distribution of antibody sequences of individuals before the vaccination and after 7 days and 4 to 5 weeks. The number of sequences analyzed for individual are shown in the inner circle. Sizes of pie slices are proportional to the number of clonally related sequences. Persisting clones (same IGV genes) in both time points are shown as colored slices. Gray indicates sequences not overlapped between individuals.
SARS-CoV-2 T-cell memory. (A) Bar graphs show the number of clonotypes in each patient after the vaccination. (B) Box plots show the induction fold of variable (V) gene usage from TCRα and TCRβ chain.
SARS-CoV-2 T-cell memory. (A) Bar graphs show the number of clonotypes in each patient after the vaccination. (B) Box plots show the induction fold of variable (V) gene usage from TCRα and TCRβ chain.
Discussion
In this study, we demonstrate that patients with CLL under active surveillance and those that are treatment-naïve, exhibit a superior response to COVID-19 vaccination than patients on active treatment. Although naïve or minimally treated patients showed an expanded humoral and cellular immune response, the heavily pretreated patients exhibited only T-cell immune responses, even with repeated immune stimulation utilizing a heterologous vaccination regimen. The value of a 3-dose vaccination was most pronounced in patients who exhibited evidence of a recuperated immune system following effective CLL treatment. These results coincided with pattern of transcriptional expression of immune genes and BCR/TCR repertoire.
Interrogation of the transcriptional response to vaccination utilizing RNA-seq highlighted that nearly all patients with CLL demonstrated transcriptional activation of early innate immune response pathways, including interferon-JAK/STAT signaling,17,25 within 2 days, regardless of the antibody response. Interferon-mediated innate immune response serves as a biomarker and plays a critical role in the immune system to control viral replication combating SARS-CoV-2 infection.17,46,47
Skewed IGHV usage, including the appearance of IGHV1-69, IGHV 4-34, and IGHV 3-21, was observed in the BCR repertoire of the patients with CLL. Diverse IGHV usage occurs in patients with COVID-19 and vaccinated individuals.17,25 In addition, the final number of clonotypic B cells detected was much lower in non-IgG responders and even in the CLL seropositive patients were lower vaccinated healthy controls. The data are consistent with increased numbers of SARS-CoV-2 specific IGHV clones correlating with an improvement of humoral response rate and B-cell reconstitution.
Hypogammaglobulinemia in patients with CLL results from leukemic cells perturbating the interaction between T and B cells. Patients with low serum immunoglobulin levels typically show ineffective humoral responses after both primary and subsequent boosting doses.14,23,48 Half of the patients with persistent immunodeficiency or B-cell depletion following initial vaccination remain seronegative after a booster dose.8,9,14,49 T-cell immunity is essential for viral recognition and clearance and cellular responses can prevent initial infection and seroconversion.10,23,49 T cells are especially critical in immune protection against SARS-CoV-2 in patients with cancer, who undergo therapy with B-cell depleting agents, such as anti-CD20 antibody.50,51 Here we report that patients with CLL with diminished numbers of functional CD19+ B cells, a key player in humoral response against SARS-CoV-2 virus, developed robust T-cell immune responses to COVID-19 vaccination.
Importantly, we also found a BTKi or BCL2i treatment-dependent effect on the immune response to COVID-19 vaccine boosters. Vaccine effectiveness is moderated by the time of vaccination in relation to CLL treatment outcomes.8,52 Here, it seems that patients with CLL treated with the BTK inhibitor ibrutinib for more than 5 years failed to seroconvert, whereas patients treated with ibrutinib within 2 years transiently seroconverted after 2 to 3 doses of a COVID-19 vaccine. Notably, seropositive patients started a frontline therapy with acalabrutinib, a second generation of BTKi, shortly before or after a 3-dose, and retained antibody response levels beyond 6 months after treatment. Prolonged BTKi treatment predisposes patients toward an ineffective vaccine immune response because B-cell maturation relies on functional BTK.53 Increased serum IgA levels in BTKi-treated patients with CLL appears to improve functional humoral immunity demonstrated by decreased infection susceptibility and hospitalization rates.2,53
Protection from severe disease, hospitalization, and death by COVID-19 vaccination results from the combination of humoral immunity with a durable cellular immune memory response. In immunocompromised patients the timing of initial and booster vaccination should be carefully considered in reference to the underlying disease status. Remission-inducing therapy resulting in improved immune status and B-cell reconstitution improves adaptive immunity.12
Limitations of the study
There are several limitations to this study. The study was conducted on volunteers from a specific geographical area, Munich, Germany, total number of patients with CLL were limited, patients received different treatment and vaccination strategies, and samples were not available from all patients for all vaccination time points for the RNA-seq studies.
Acknowledgments
The authors thank the participants who contributed to this study to advance their understanding of COVID-19 vaccination. This work used the computational resources of the National Institutes of Health (NIH) High Performing Computation Biowulf cluster (http://hpc.nih.gov). RNA-sequencing was conducted in the NIH Intramural Sequencing Center (https://www.nisc.nih.gov/contact.htm).
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, US National Institutes of Health, and the Bavarian State Ministry for Science and Art, Special Program for the Promotion of COVID-19 Research, Germany (AKZ H.40001.1.7 [M.B.]).
Authorship
Contribution: H.K.L., M.A.H., P.A.F., C.-M.W., and L.H. designed the study; H.K.L. performed RNA sequencing and data analysis, and prepared the figures; M.A.H. and T.T.P. selected the patients and collected and curated data; H.K.L., M.A.H., M.B., T.T.P., P.A.F., C.-M.W., and L.H. analyzed results; H.K.L. and T.T.P. prepared the table; M.A.H., M.B., T.T.P., J.W.H., K.M., S.Z., H.v.B., P.G., R.W., L.B., and L.P. prepared samples; H.K.L., M.A.H., T.T.P., M.B., P.A.F., C.-M.W., and L.H. wrote the manuscript; and all authors read and approved the final version of the manuscript.
Conflict-of-interest disclosure: M.A.H. received consultancy fees from AstraZeneca. C.M.W. received consultancy fees from AstraZeneca and BioNTech. The remaining authors declare no competing financial interests.
Correspondence: Lothar Hennighausen, National Institutes of Health, 8 Center Dr, Bethesda, MD 20892; e-mail: lotharh@nih.gov; and Clemens-Martin Wendtner, Munich Clinic, Kölner Platz1, 80804 München, Germany; e-mail: clemens.wendtner@muenchen-klinik.de.
References
Author notes
The RNA-seq data reported in this article have been deposited in the Gene Expression Omnibus database (GEO) (accession number GSE201642) and is publicly available (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201642). RNA-seq data of healthy heterologous vaccinated individuals were obtained under GSE201535 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201535).
Data are available on request from the corresponding authors, Lothar Hennighausen (lotharh@nih.gov) and Clemens-Martin Wendtner (clemens.wendtner@muenchen-klinik.de).
The full-text version of this article contains a data supplement.
H.K.L., M.A.H., and M.B. are joint first authors.
C.M.W. and L.H. are joint senior authors.

![Study population and design. (A) CLL patient groups by treatment (naïve [n = 2], treatment active [n = 7], and off-treatment [n = 6]) and by drugs (BTKi [n = 3], Bcl2i [n = 4], anti-CD20 for less than 12 months [n = 4], and anti-CD20 for more than 12 months [n = 2]). (B) Blood samples were collected before the second (II-D0) and third (III-D0) vaccination and at days 2 (D2), 7 (D7) and 14 (D14) and weeks 4 to 5 (W4-5) after second and third vaccination as indicated by the colored circles. Mean and standard deviation for the number of days between first and second vaccinations was 66 ± 25 days and between second and third vaccinations was 131 ± 43 days. (C) The number of patients who received each vaccine regimen.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/10/10.1182_bloodadvances.2022008445/2/m_blooda_adv-2022-008445-gr1.jpeg?Expires=1767703808&Signature=gQjJI31kS34rDkt2s4XI75p9-QKpRh3bjGZV39KAbIGtJ2pUu2iEOItS6Xf99vysGz933VEbQAHIc2SVroiZ3drRYFI8Uhh-PPjRPcwBNT3jFadtOvKBcfgNNJamy~X3z3mX8YoJOZWg~aPffPEpa0d9GJ3YhZwkzfydZsDVeVK5Te2n8qacG1mlh5d7U~Pe4vxJkn9SJGj3gdWvAbPF6zwWftZfRa0LiEJO3RdMCtt3XyAHiiHFXJWo7-BnM6QdJDsqtn5JJL0iMElcygmKGD37AQkNe1ArZa~iIvHbryroiveksm3gXbavmVlH0J1o~sbuHpr6OLVomauNrRm~Aw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)