Key Points
TFPI-binding exosite mutations have been reported in patients with FVII deficiency and clinical bleeding variabilities.
The alteration of TFPI-binding exosites of FVII affected FVII secretion and the binding with FX, resulting in abnormal bleeding.
Abstract
Tissue factor (TF) pathway inhibitor (TFPI) is a Kunitz-type anticoagulation protein that inhibits activated factor VII (FVIIa)/TF complex. Incidentally, many different F7 gene variants, including TFPI-binding exosite mutations, have been reported in patients with congenital FVII deficiency and clinical bleeding variabilities. Here, TFPI-binding exosites (R147 and K192) on FVII zymogen were selectively disrupted to understand their roles in the pathogenesis of bleeding phenotypes. Expression of recombinant FVII variants (R147A, K192A, and R147A/K192A) demonstrated markedly reduced secretion of FVII owing to intracellular retention in the endoplasmic reticulum, as demonstrated by upregulation of the unfolded protein response genes in all FVII variants. FVII variants showed a similar FVII activation pattern and FVIIa amidolytic activity than FVII wild-type (WT). In contrast to FVII activation, R147A and K192A showed a 90% reduction in FX activation relative to WT, whereas the R147A/K192A variant demonstrated a 99% decrease in FX activation. The clotting time was markedly prolonged with R147A and K192A than WT, and no FVII coagulant activity was detected in R147A/K192A. In addition, the thrombin generation assay revealed a significant prolongation of lag time in all FVII variants. Our study explains how mutations of TFPI-binding exosites of FVII can lead to bleeding phenotypes in individuals carrying these aberrancies.
Introduction
Factor VII (FVII) is a vitamin K–dependent coagulation factor that plays a pivotal role in the initiation phase of the coagulation system.1 After vascular injury, a local hemostasis is initiated when an injured vessel wall exposes tissue factor (TF) to the blood circulation. FVII binds to TF and is converted to its activated form (FVIIa). The FVIIa/TF complex subsequently activates FIX and FX, leading to a burst of thrombin generation that causes fibrin clot formation.2,3
Tissue factor pathway inhibitor (TFPI) is a Kunitz-type anticoagulation protein produced by endothelial cells.4 It is released almost immediately after the initiation phase of coagulation. TFPI acts as a major physiological inhibitor of the extrinsic pathway by binding to TF/FVIIa/FXa, forming a quaternary complex.5 This binding relies on the interaction between the amino acids D11, R20, and E46 (chymotrypsin numbering system) of the Kunitz-1 domain of TFPI and R147/K192, D60, and K60a/K60c of FVIIa, respectively.4,6
Given that TFPI is responsible for FVIIa inhibition, mutations on TFPI-binding exosites of FVII should disrupt the interaction between FVII and TFPI, hence inhibiting the effective binding of the 2 proteins. Ideally, FVII with TFPI-binding exosite mutations should demonstrate the prolongation of plasma half-life compared with wild-type (WT) FVII, and individuals carrying mutations at these exosites should be at high risk for thromboembolic phenomena. In contrast, many different F7 gene variants, including TFPI-binding site mutations that cause bleeding disorders with clinical variability, have been reported. Particular mutations (IVS6+1G>T, K376X, IVS2+2T>C) were strongly associated with intracranial hemorrhage.7 TFPI-binding site mutations R147C and K192E were reported in patients with reduced FVII activity.8-10 Moreover, the R147C mutation was detected in 2 patients with severe bleeding symptoms, namely hemarthrosis, easy bruising, and hematoma.10 In addition, compound heterozygous mutations K192E along with IVS6-1G>A or nt27 del CT mutations were detected with prolonged prothrombin time (PT) and reduced FVII activity and FVII antigen.8,9 However, the underlying molecular mechanisms of these mutations have not been thoroughly investigated. To understand the pathogenicity of previously reported mutations of FVII, the molecular pathogenic mechanism of these point mutations on FVII protein should be characterized for diagnostic and prognostic use.
In this study, we investigated the impact of altering TFPI-binding exosites R147 and K192 of FVII on the structural and functional properties of the FVII protein to understand their role in the pathogenesis of bleeding disorders involving FVII mutations.
Methods
DNA mutagenesis and plasmid constructions
pTG19-T PCR cloning vector (Vivantis Technologies, Sdn Bhd, Malaysia) containing human FVII complementary DNA (cDNA) was used as a template for mutant FVII cloning. Alanine substitutions were introduced into the F7 sequence by polymerase chain reaction (PCR)–based mutagenesis. Mutagenesis primers are listed in Table 1. The constructs containing R147A and K192A were verified by full-length sequencing of the cDNA (Macrogen Inc, Seoul, Korea). The correct clone was cut with XhoI and BamHI. The PCR product size of FVII with 1357 bp was confirmed on an agarose gel stained with 1% GelRed (Biotium, Hayward, CA) and purified using a GenepHlow gel/PCR kit (Geneaid Biotech, New Taipei, Taiwan). The purified PCR product was ligated into the pLVX-puro vector (Clonetech Laboratories, Mountain View, CA), which was further confirmed by sequencing. This construct was used as a template for FVII mutant expression.
Primers used for generating the FVII mutations
| Mutation . | Primer . |
|---|---|
| R147A forward: | 5′- CAG CTG CTG GAC GCC GGC GCC ACG -3′ |
| R147A reverse: | 5′- CAG GGC CGT GGC GCC GGC GTC CAG -3′ |
| K192A forward: | 5′- G GAC TCC TGC GCC GGG GAC AGT GGA G -3′ |
| K192A reverse: | 5′- C TGT CCC CGG CGC AGG AGT CCT TGC TGC -3′ |
| Mutation . | Primer . |
|---|---|
| R147A forward: | 5′- CAG CTG CTG GAC GCC GGC GCC ACG -3′ |
| R147A reverse: | 5′- CAG GGC CGT GGC GCC GGC GTC CAG -3′ |
| K192A forward: | 5′- G GAC TCC TGC GCC GGG GAC AGT GGA G -3′ |
| K192A reverse: | 5′- C TGT CCC CGG CGC AGG AGT CCT TGC TGC -3′ |
Codon for mutated amino acid is in bold.
Cell culture condition
Human embryonic kidney 293T (HEK293T) cells were used for stable transfection and FVII protein expression. HEK293T cells were grown in DMEM/high-glucose medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (FBS; Gibco, Corning, NY), 2 mM L-glutamine (HyClone), 100 U/mL penicillin, and 100 μg/mL streptomycin (HyClone). Cells were maintained in 37°C incubator with 5% CO2 and 95% humidity.
Expression of FVII molecules in mammalian cells
HEK293T cell lines expressing human FVII WT, R147A, K192A, and R147A/K192A were constructed via stable transfection with WT and mutant plasmids using FuGENE6 transfection reagent (Promega, Madison, WI) according to the manufacturer’s protocol. Transfected cells were selected after growth in a complete medium containing 2 μg/mL puromycin for 14 days. Puromycin-resistant clones were chosen and tested for FVII protein expression in cell culture supernatants by western blot analysis.
After the stable clones of FVII were successfully established, the transfected cells were cultured in serum-free media supplemented with 1× insulin-transferrin-selenium and 10 μg/mL vitamin K for 24 hours at 37°C. Cell culture supernatants were collected and supplemented with benzamidine, CaCl2, and PEG-8000 at the final concentration of 10 mM, 5 mM, and 0.1%, respectively. The collected media were centrifuged at 3000 × g for 10 minutes at 4°C to remove cell debris and then stored in aliquots at −80°C. The transfected cells were washed 3 times with ice-cold 1× phosphate-buffered saline (PBS). These cells were lysed in lysis buffer (Bio-Rad, Berkeley, CA) containing protease inhibitor cocktail (Bio-Rad). FVII protein levels were measured in both cell lysates and culture supernatants using western blotting and enzyme-linked immunosorbent assay (ELISA), respectively.
SDS-PAGE and western blot analysis
The expression of FVII protein was analyzed by western blotting. Protein samples were linearized by mixing with disulfide-reducing Laemmli buffer (Bio-Rad), boiled at 95°C for 5 minutes, and subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred from the gel to the PVDF membrane (Thermo Fisher Scientific, Waltham, MA). The blot was blocked with 5% skimmed milk in TBS-T (10 mM Tris, 150 mM sodium chloride, 0.05% Tween 20, pH 7.4), probed with the monoclonal rabbit anti-FVII at 1:10 000 dilution (Abcam, Cambridge, United Kingdom) for 3 hours at room temperature (RT), and incubated with peroxidase-conjugated goat anti-rabbit IgG H&L at 1:20 000 dilution (Abcam) for 1 hour at RT. Immunoreactive proteins were detected by using ECL detection reagents (Bio-Rad). The membrane was then exposed to the gel documentation system for chemiluminescence imaging.
Measurement of FVII protein levels in culture media
The amount of secreted FVII in the culture medium was measured by ELISA using the Human Factor VII ELISA Kit (Abcam) according to the manufacturer’s instructions. A standard curve was established by twofold serial dilutions to produce 8 standard points of human FVII standard protein (0-200 ng/mL). Three independent experiments in duplicates were performed. The absorbance was read in a Varioskan Flash microplate reader (Thermo Fisher Scientific) at a wavelength of 450 nm and compared with a standard curve.
Confocal immunofluorescence microscopy
The transfected cells (2 × 105 cells per well) were plated on coverslips precoated with poly-D-lysine in a 6-well plate. After 24 hours of seeding, cells were washed with 1× PBS and fixed with 4% paraformaldehyde for 10 minutes at RT. Cells were washed 3 times with PBS and blocked in blocking solution (89.9% 1× PBS, 10% FBS, 0.1% Triton X-100) for 30 minutes at RT. Immunostaining was performed by incubating with primary antibodies: rabbit monoclonal anti-FVII at 1:500 dilution (Abcam), mouse monoclonal anti–protein disulfide isomerase (PDI) at 1:100 dilution (Thermo Fisher Scientific), or mouse polyclonal anti-GM130 at 1:250 dilution (Abcam) diluted in antibody solvent solution (98.9% 1× PBS, 1% FBS, 0.1% Triton-X100) for 2 hours at RT. Cells were washed twice with 1× PBS. Cells were incubated with secondary antibodies: Alexa Fluor 488 goat anti-rabbit at 1:1000 dilution (Abcam) and Cy3 goat anti-mouse at 1:500 dilution (Abcam) for 1 hour at RT. Cells were washed 3 times with 1× PBS and mounted in the mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Abcam). Cells were observed and imaged using the Eclipse Ts2R inverted research microscope (Nikon Instruments Inc, Melville, NY) and Nikon NIS-Elements D software. FVII colocalizations with the endoplasmic reticulum (ER) or the Golgi apparatus were statistically evaluated using Pearson’s correlation coefficient (PCC), relying on JacoP plugin for the ImageJ software program.
Quantitative reverse transcription PCR (qRT-PCR) analysis
Total RNA was extracted from transfected cells by FavorPrep Blood/Cultured Cell Total RNA Purification Mini Kit (Favorgen, Ping-Tung, Taiwan) following the manufacturer’s instructions. One microgram of total RNA was converted into cDNA using iScript Reverse Transcription Supermix (Bio-Rad). To measure messenger RNA (mRNA) levels of various ER stress–related genes, qRT-PCR was performed using KAPA SYBR FAST qPCR master mix (2×) kit (KAPA Biosystems, Wilmington, MA). Gene expression profiles were determined on an ABI7500 real-time PCR system (Applied Biosystems). The specific primers of ER stress–related genes (ATF4, BiP, CHOP, XBP1, and ATF6) used here were those used in a previous study.11 The cycling conditions consisted of 3 minutes at 95°C, followed by 40 cycles at 95°C for 10 seconds, 61°C for 20 seconds, and 72°C for 30 seconds. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. The data were presented as a relative gene expression level by using the 2−ΔΔCt method.
Determination of FVII coagulant activity
FVII activation
FVII activation was determined in a reaction containing 0.5-nM recombinant FVII (rFVII) and 60-nM FXa (Abcam) at 37°C. The reaction was sampled at 0 and 30 minutes to determine FVII activation by western blotting as described above.
FVIIa amidolytic activity
FVII (15 nM) was activated by 60-nM FXa in a reaction buffer (20-mM Hepes/150-mM sodium chloride, pH 7.4) at 37°C for 30 minutes. Rivaroxaban (120 nM; Bayer Pharma AG, Leverkusen, Germany) was then added to inhibit the activity of FXa at 37°C for 30 minutes. FVIIa substrate (0.8 mM; Sigma-Aldrich, St. Louis, MO) was added immediately before monitoring absorbance at 405 nm using a Varioskan Flash microplate reader (Thermo Fisher Scientific). The actual absorbance for FVIIa-specific amidolytic activity was calculated by ODtotal reaction − ODFXa+rivaroxaban.
FX activation by FVII variants with TF
The abilities of FVII variants to activate FX to FXa were examined using the Factor VII Human Chromogenic Activity Kit (Abcam) according to the manufacturer’s instructions. FVII from each variant was diluted to 500 pM (25 ng/mL) in sample diluents and run in duplicate. A standard curve was established by twofold serial dilutions to produce 6 standard points of human FVII standard protein (0-0.2 IU/mL). The absorbance was read in a Varioskan Flash microplate reader (Thermo Fisher Scientific) at 405 nm every 30 seconds for 30 minutes and compared with the standard curve.
Thrombin generation assay
Thrombin generation in FVII-deficient plasma was measured using the calibrated automated thrombogram method, which cleaves a fluorogenic substrate (Z-Gly-Gly-Arg-AMC) to continuously measure thrombin generation in clotting plasma. A calibration curve was constructed in the same FVII-deficient plasma with a fixed amount of thrombin–α2-macroglobulin complex (thrombin calibrator; Thrombinoscope B.V., Maastricht, The Netherlands). The ability of FVII variants to support TF-triggered thrombin generation was determined by adding 20 μL of a mixture of 0.2-pM TF and 2-μM phospholipids (PPP Reagent LOW; Stago, Parsippany-Troy Hills, NJ) to 70 μL of FVII-deficient plasma (Siemens, Midrand, South Africa) and 10 μL of 7-nM FVII variants, with preheating at 37°C for 10 minutes. Assays were initiated with 20 μL of fluorogenic substrate and calcium solution (FluCal; Stago). Fluorescence was read in a Fluoroskan Ascent reader (Thermo Labsystems OY, Helsinki, Finland) and thrombin generation over time was calculated using the Thrombinoscope software (Thrombinoscope B.V.). Four parameters were derived from the thrombin generation curves: peak thrombin (nM), endogenous thrombin potential (ETP) (nM·min), lag time (minutes), and time to peak (minutes).
Clotting assay
The abilities of FVII variants on coagulant activity were examined by a modified PT assay using lyophilized human placental thromboplastin, Thromborel S (Siemens) as the activator in lyophilized human FVII–deficient plasma (Siemens). FVII samples (5 nM) were added to FVII-deficient plasma. The reaction was initiated by the addition of Thromborel S reagent into the mixture. The time to clot formation was recorded on the semiautomated coagulation analyzer CA-104 (Sysmex Europe GmbH, Norderstedt, Germany).
Statistical analysis
Data were expressed as mean ± standard error of the mean (SEM) values (n ≥ 3), and the mean levels in different groups were compared using one-way analysis of variance (ANOVA) followed by a Tukey multiple comparison test. P < .05 was considered statistically significant. The figures and all statistical analyses were prepared using GraphPad Prism 5.0 (GraphPad Software Inc, La Jolla, CA).
Results
Cells transfected with FVII expressed the zymogen form of FVII
Alanine substitutions were introduced into the TFPI-binding exosites (R147 and K192) on the FVII protease domain. After sequencing, R147A, K192A, and R147A/K192A variants demonstrated successful alanine substitution. Arginine (R) at position 147 and lysine (K) at position 192 were changed to GCC sequences, resulting in alanine substitution, which was confirmed by DNA sequencing chromatogram (Figure 1A,C) and DNA sequence alignment with WT and primer (Figure 1B,D). After transfection, the forms of secreted FVII proteins in the culture media supernatant were revealed by western blotting. FVII was not detectable in nontransfected FVII cells. All HEK293T transfected with FVII constructs (WT, R147A, K192A, and R147A/K192A) expressed the zymogen form of FVII as a single band corresponding to a molecular weight of 55 kDa (Figure 1E). FVIIa showed the light chain of protease domain (MW ∼ 20 kDa).
Constructionand expressionof human FVII. DNA sequencing chromatogram of R147A (A) and K192A (C). After sequencing, R147A, K192A, and R147A/K192A variants demonstrated the successful alanine substitution that changed nucleotide sequences at position 147 and 192 to GCC, resulting in alanine substitution. The red underline represents GCC sequences. The green color represents nucleotide A. The blue color represents nucleotide C. The black color represents nucleotide G. The red color represents nucleotide T. Sequence alignment of R147A (B) and K192A (D) with WT FVII and primer. The codon at position R147 and K192 was similar to their primers. The star represents similar sequences. Western blot analysis of FVII variants in culture media supernatant (E). One microgram of total protein was applied to each lane. Nontransfected HEK (1 μg of total protein) was used as a negative control. Recombinant FVIIa (NovoSeven, 100 ng) was used as a positive control. The zymogen FVII and the light chain of the protease FVIIa appeared as a single band at 55 and 20 kDa, respectively. Secreted FVII protein levels in culture media supernatant (F). FVII protein levels were determined by ELISA. FVII mRNA levels of FVII variants (n = 3) (G). FVII mRNA levels were examined by qRT-PCR. Intracellular FVII proteins. After stable transfection, FVII protein levels in cell lysates were determined using ELISA (H). Data were reported as mean ± SEM (n = 3). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 in comparison between WT and each FVII variant.
Constructionand expressionof human FVII. DNA sequencing chromatogram of R147A (A) and K192A (C). After sequencing, R147A, K192A, and R147A/K192A variants demonstrated the successful alanine substitution that changed nucleotide sequences at position 147 and 192 to GCC, resulting in alanine substitution. The red underline represents GCC sequences. The green color represents nucleotide A. The blue color represents nucleotide C. The black color represents nucleotide G. The red color represents nucleotide T. Sequence alignment of R147A (B) and K192A (D) with WT FVII and primer. The codon at position R147 and K192 was similar to their primers. The star represents similar sequences. Western blot analysis of FVII variants in culture media supernatant (E). One microgram of total protein was applied to each lane. Nontransfected HEK (1 μg of total protein) was used as a negative control. Recombinant FVIIa (NovoSeven, 100 ng) was used as a positive control. The zymogen FVII and the light chain of the protease FVIIa appeared as a single band at 55 and 20 kDa, respectively. Secreted FVII protein levels in culture media supernatant (F). FVII protein levels were determined by ELISA. FVII mRNA levels of FVII variants (n = 3) (G). FVII mRNA levels were examined by qRT-PCR. Intracellular FVII proteins. After stable transfection, FVII protein levels in cell lysates were determined using ELISA (H). Data were reported as mean ± SEM (n = 3). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 in comparison between WT and each FVII variant.
Secreted FVII levels were reduced in cells expressing FVII variants
The secreted FVII levels in culture supernatant from the transfected HEK293T cell lines were examined using ELISA. Among the HEK293T cell lines transfected with the FVII constructs, WT secreted the highest amount of FVII proteins (3031.56 ± 93.06 ng/mL) than other FVII variants. FVII levels in the single-mutant R174A and K192A were 1249.37 ± 129.56 ng/mL and 1266.41 ± 57.73 ng/mL, respectively. FVII level in the double-mutant R147A/192A was 1018.62 ± 128.72 ng/mL (Figure 1F). This result demonstrated that FVII variants significantly decreased FVII protein production compared with WT in the transfected HEK293T. To illustrate whether the decreased FVII protein expression of FVII variants was due to the differences in their mRNA levels, the mRNA expression levels of WT and FVII variants were determined by qRT-PCR in stable transfected cells. No differences in the FVII RNA levels were observed between WT and FVII variants in transfected cells (Figure 1G).
FVII variants were retained in the ER
Because the FVII variants (R147A, K192A, and R147A/K192A) demonstrated significantly reduced secretion levels, we postulated that FVII variants could be intracellularly retained. Therefore, FVII proteins in the cell lysates and intracellular localization of the FVII variants in transfected HEK293T cells were determined by western blotting and immunofluorescence staining, respectively. The intracellular levels of FVII R147A, K192A, and R147A/K192A determined by ELISA showed a significant increase (3770.42 ± 235.10, 3555.52 ± 178.04, and 3283.64 ± 73.70 ng/mL, respectively) compared with WT (2468.96 ± 71.75 ng/mL) (Figure 1H). The intracellular localization study demonstrated that both WT and FVII variants colocalized with the ER marker PDI (Figure 2A) and the Golgi marker GM130 (Figure 2C). However, the FVII variants (R147A, K192A, and R147A/K192A) predominately colocalized with PDI compared with WT (Figure 2A). The PCC measurements in the ER and the Golgi apparatus were calculated to quantify FVII colocalization with subcellular organelles. The PCC value in the ER was significantly increased in the FVII variants (Figure 2B), whereas the PCC value in the Golgi was significantly decreased in the FVII variants compared with WT (Figure 2D). These results supported the immunofluorescence findings and indicated that the mutant FVII proteins were primarily retained in the ER and led to an impaired transport of the FVII proteins from the ER to the Golgi.
Intracellular localization of FVII variants. Intracellular localization of FVII variants in the ER (A) and the Golgi (B). The markers of cell organelles (ER and Golgi) were stained in red color. FVII was stained in green. Colocalization of FVII with a cell organelle marker (ER or Golgi) was shown in yellow color. Scale bars, 10 μm. The colocalization of FVII with PDI (C) and GM130 (D) was calculated by the PCC. The PCC score was estimated from 10 cells for each FVII. ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 in comparison between WT and each FVII variant.
Intracellular localization of FVII variants. Intracellular localization of FVII variants in the ER (A) and the Golgi (B). The markers of cell organelles (ER and Golgi) were stained in red color. FVII was stained in green. Colocalization of FVII with a cell organelle marker (ER or Golgi) was shown in yellow color. Scale bars, 10 μm. The colocalization of FVII with PDI (C) and GM130 (D) was calculated by the PCC. The PCC score was estimated from 10 cells for each FVII. ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 in comparison between WT and each FVII variant.
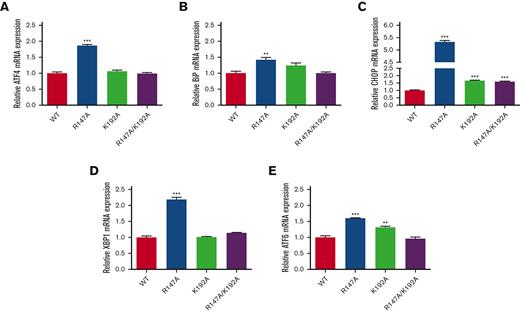
ER stress–related genes were upregulated in cells expressing FVII variants
To determine whether FVII variants induced ER stress in transfected HEK293T cells, the transcription levels of the downstream target genes of the unfolded protein response (UPR) pathway in cells expressing FVII variants, consisting of ATF4, ATF6, BiP, CHOP, and XBP1 were examined by qRT-PCR. The mRNA expression levels of all ER stress–related genes were higher in all cells expressing FVII variants (R147A, K192A, and R147A/K192A) than in cells expressing WT (Figure 3). Interestingly, the highest mRNA levels of all ER stress–related genes (ATF4, BiP, CHOP, XBP1, and ATF6) were observed in R147A-expressing cells by 1.87 ± 0.03, 1.42 ± 0.07, 5.33 ± 0.05, 2.19 ± 0.07, and 1.60 ± 0.01 folds, respectively (Figure 3). The data indicated that FVII variants caused ER stress in the FVII variant–transfected HEK293T cells.
Expression of ER stress–related genes. ATF4 (A), BiP (B), CHOP (C), XBP1 (D), and ATF6 (E) were determined in each FVII variant–transfected HEK293T by qRT-PCR. The results were presented as mean ± SEM (n = 3). ∗∗∗P < .001; ∗∗P < .01 in comparison between WT and each FVII variant.
Expression of ER stress–related genes. ATF4 (A), BiP (B), CHOP (C), XBP1 (D), and ATF6 (E) were determined in each FVII variant–transfected HEK293T by qRT-PCR. The results were presented as mean ± SEM (n = 3). ∗∗∗P < .001; ∗∗P < .01 in comparison between WT and each FVII variant.
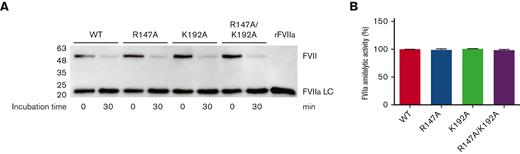
FVII variants were normally activated to FVIIa by FXa
FVII activation was monitored by incubating FVII variant (0.5 nM) with FXa (60 nM) for 30 minutes at 37°C. Under these conditions, all FVII constructs (WT, R147A, K192A, and R147A/K192A) were immediately activated to FVIIa, as demonstrated by the appearance of bands corresponding with the light chain of protease FVIIa (MW ∼ 20 kDa) at 0 minute (Figure 4A). After the 30-minute incubation, all variants were more activated to FVIIa by increasing band intensity corresponding with the light chain of protease FVIIa and decreasing band intensity corresponding with the zymogen FVII (Figure 4A). After the 60- and 120-minute incubation, all variants demonstrated band patterns similar to those after the 30-minute incubation (data not shown). Therefore, the 30-minute incubation was used to activate FVII to determine FVIIa amidolytic activity toward a peptidyl substrate FVIIa.
FVII activation by FXa determined by western blotting and FVIIa amidolytic activity. (A) Equivalent molar concentrations (0.5 nM) of WT, R147A, K192A, and R147A/K192A were incubated with 60-nM FXa for 0 and 30 minutes at 37°C. At each time point, the enzymatic reactions were quenched with the reducing sample buffer, run on 12% SDS-PAGE, transferred to PVDF membrane, and probed with the rabbit anti–FVII monoclonal antibody as described in the Methods section. FVIIa LC, the light chain of protease FVIIa; rFVIIa, recombinant FVIIa (NovoSeven, 10 ng). (B) FVII (15 nM) was activated by FXa in reaction buffer at 37°C in the presence of 60-nM FXa for 30 minutes. Rivaroxaban (120 nM) was then added to the inhibit activity of FXa at 37°C for 30 minutes. FVIIa substrate (0.8 mM) was added to determine FVIIa amidolytic activity. Data were presented in percentage of activity compared with WT FVII.
FVII activation by FXa determined by western blotting and FVIIa amidolytic activity. (A) Equivalent molar concentrations (0.5 nM) of WT, R147A, K192A, and R147A/K192A were incubated with 60-nM FXa for 0 and 30 minutes at 37°C. At each time point, the enzymatic reactions were quenched with the reducing sample buffer, run on 12% SDS-PAGE, transferred to PVDF membrane, and probed with the rabbit anti–FVII monoclonal antibody as described in the Methods section. FVIIa LC, the light chain of protease FVIIa; rFVIIa, recombinant FVIIa (NovoSeven, 10 ng). (B) FVII (15 nM) was activated by FXa in reaction buffer at 37°C in the presence of 60-nM FXa for 30 minutes. Rivaroxaban (120 nM) was then added to the inhibit activity of FXa at 37°C for 30 minutes. FVIIa substrate (0.8 mM) was added to determine FVIIa amidolytic activity. Data were presented in percentage of activity compared with WT FVII.
FVII variants can cleave a peptidyl substrate
After successful FXa-mediated FVII activation, FVIIa-specific amidolytic activity was determined using a chromogenic assay toward the hydrolysis of peptidyl substrate FVIIa. Recombinant WT and all FVII variants (R147A, K192A, and R147A/K192A) demonstrated similar amidolytic activity (Figure 4B).
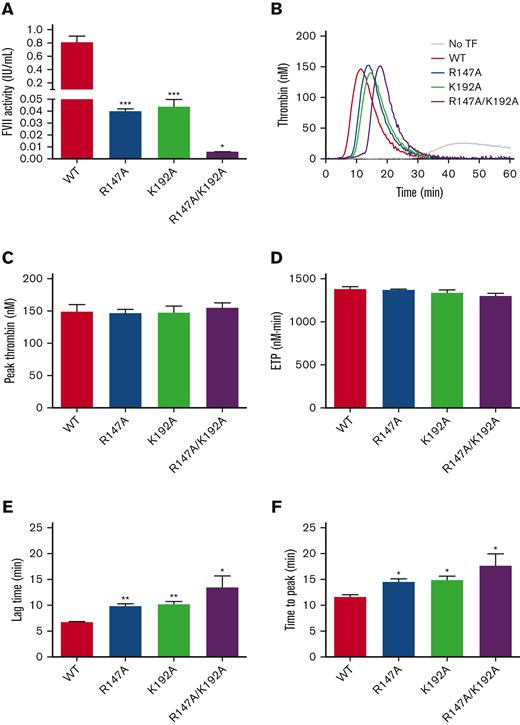
FVII variants demonstrated the decreased ability to activate FX
FX activation by FVII variants was measured using a 2-step FXa generation assay. WT demonstrated the highest FVII activity (0.8090 ± 0.0948 IU/mL). R147A and K192A showed mildly reduced FX activation than WT (0.0399 ± 0.0018 IU/mL and 0.0437 ± 0.0062 IU/mL, respectively). The R147A/K192A demonstrated a markedly reduced FX activation relative to WT (0.006 ± 0.00006 IU/mL) (Figure 5A).
FVII-mediated FX activation determined by chromogenic assay (2-step FXa generation) and the ability of FVII variants to support plasma thrombin generation in FVII-deficient plasma. (A) FVII from each variant was diluted to 500 pM and used to evaluate coagulant activity. Data were reported as FVII activity (mean ± SEM, n = 3). (B) Representative thrombin generation curves showing the ability of FVII variants to support TF-triggered thrombin generation. (C) The mean peak thrombin and (D) ETP values of all FVII variants were similar to those of WT. (E) Lag time and (F) time-to-peak values of FVII variants were significantly longer than those of WT. The results were presented as mean ± SEM (n = 3). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 in comparison between WT and each FVII variant.
FVII-mediated FX activation determined by chromogenic assay (2-step FXa generation) and the ability of FVII variants to support plasma thrombin generation in FVII-deficient plasma. (A) FVII from each variant was diluted to 500 pM and used to evaluate coagulant activity. Data were reported as FVII activity (mean ± SEM, n = 3). (B) Representative thrombin generation curves showing the ability of FVII variants to support TF-triggered thrombin generation. (C) The mean peak thrombin and (D) ETP values of all FVII variants were similar to those of WT. (E) Lag time and (F) time-to-peak values of FVII variants were significantly longer than those of WT. The results were presented as mean ± SEM (n = 3). ∗∗∗P < .001; ∗∗P < .01; ∗P < .05 in comparison between WT and each FVII variant.
FVII variants delayed plasma thrombin generation activity
The ability of FVII variants to support TF-triggered thrombin generation was examined in FVII-deficient plasma supplemented with 1-nM FVII variants (Figure 5B). The mean peak thrombin (Figure 5C) and ETP (Figure 5D) values of all FVII variants were similar compared with those of WT. FVII variants (R147A, K192A, and R147A/K192A) demonstrated significantly longer lag time (Figure 5E) and time-to-peak (Figure 5F) values than WT.
Plasma-based coagulant activities of FVII variants were decreased than those of WT
Modified PT assay was used to examine the ability of FVII variants to generate fibrin clots in plasma. WT completely normalized the clotting time (18.60 ± 0.39 seconds), whereas the clotting time was significantly prolonged with R147A and K192A. The clotting time of R147A/K192A was markedly prolonged to 95.13 ± 0.99 seconds. Such prolonged clotting time was not in the range of standard curve, and the activity could not be determined (Table 2).
Coagulant activity of FVII variants determined by a modified PT assay
| rFVII . | Clotting time ± SEM, s . | FVII activity, ng/mL . |
|---|---|---|
| WT | 18.60 ± 0.39 | 88.04 ± 2.92 |
| R147A | 48.82 ± 1.65 | 6.91 ± 0.93 |
| K192A | 58.22 ± 1.81 | 3.12 ± 0.45 |
| R147A/K192A | 95.13 ± 0.99 | ND |
| rFVII . | Clotting time ± SEM, s . | FVII activity, ng/mL . |
|---|---|---|
| WT | 18.60 ± 0.39 | 88.04 ± 2.92 |
| R147A | 48.82 ± 1.65 | 6.91 ± 0.93 |
| K192A | 58.22 ± 1.81 | 3.12 ± 0.45 |
| R147A/K192A | 95.13 ± 0.99 | ND |
After incubation of FVII variants (5 nM) with FVII-deficient plasma for 5 minutes, coagulant activity was determined using a modified PT assay. Clotting times were converted to FVII levels using a FVII standard curve. Data were expressed as mean ± SEM (n = 3).
ND, not detectable.
Discussion
In this study, we examined the pathogenic mechanism underlying bleeding phenotype in FVII deficiency caused by the TFPI-binding site mutations R147 and K192 on FVII by stable transfection in HEK293T cells with cDNAs encoding the WT, R147A, K192A, and R147A/K192A variants. These mutations (R147A, K192A, and R147A/K192A) led to reduced secretion of the FVII variants relative to the WT. The decreased levels of secreted FVII variants in our study were in good agreement with the observed plasma FVII protein levels in patients with FVII deficiency carrying the mutation at K192 (K192E) who had a reduction in FVII antigen (FVII:Ag) and FVII activity (FVII:C) in the range of 38% to 44% and 2% to 3%, respectively.8,9
Amino acid substitutions affect the 3-dimensional structure of proteins that can potentially cause protein unfolding and misfolding, leading to diseases.12,13 Moreover, it was revealed that the impaired secretion of mutated coagulant proteins in some coagulation factor deficiencies was caused by ER retention.14-17 Here, immunofluorescence staining demonstrated that the FVII variants were predominantly retained in the ER (Figure 4A) and transported to the Golgi inefficiently compared with WT, indicating a misfolding of the mutant proteins affecting their normal trafficking.18 Accumulations of unfolded or misfolded proteins within the ER resulted in ER stress and induced the UPR.19 The ER stress response is mediated by 3 major receptors: IRE1, PERK, and ATF6 located in the ER membrane.20 The IRE1, PERK, and ATF6 pathways are activated by the release of BiP, which is a sensor for unfolded proteins in the ER.21 BiP expression is increased after ATF6 and IRE1 activation.22 The PERK pathway activates transcription genes involved in the ER stress–induced apoptosis such as ATF4 and CHOP.23 In the IRE1 pathway, the release of BiP from the IRE1 receptor activates XBP1 that induces the transcription of genes involved in protein folding and degradation.24 We observed the highest upregulation of all ER stress–related genes in cells expressing R147A compared with WT. The expression of CHOP was upregulated in K192A and the R147A/192A-expressing cells. Moreover, we found that ATF6 mRNA was significantly upregulated in K192A-expressing cells. Mutations disrupt protein folding in the ER and also conformational change of a protein.25,26 Therefore, proteins that cannot adopt their native conformation are retained in the ER.26 It can be observed in several conformational diseases that are caused by mutations altering the folding pathway of secretory proteins, such as diabetes and Alzheimer disease.27,28 Our results showed that the K192A and R147A/192A-expressing cells did not have much ER stress compared with the R147A-expressing cells. The alanine substitution at position R147 may greatly alter the native conformation of the protein, leading to the disruption of protein folding, causing the ER stress and activation of the UPR as presented by elevated level of all ER stress–related genes in cells expressing R147A. In contrast, the alanine substitution at position K192 may have a slight effect on the conformational change of the protein. In addition, we postulated that there were compensatory chemical bonds occurring in the molecule of R147A/K192A that might prevent protein misfolding, resulting in a slight activation of ER stress compared with R147A-expressing cells. Taken together, these results indicate that the reduced secretion of the FVII variants was due to the intracellular retention that subsequently induced the ER stress and upregulation of the UPR genes in all FVII variants.
In addition to the protein retention, we further evaluated the enzymatic and coagulant activities of FVII variants. Activations of all FVII variants by FXa appeared intact and demonstrated a time-dependent fashion, which was similar to those observed in the WT. Moreover, all FVII variants showed similar FVIIa amidolytic activity to WT. Our result was consistent with a previous study showing that the amidolytic activity of R147A in the presence of TF using 3 different peptidyl p-nitroaniline substrates was indistinguishable from the activity of WT FVIIa, indicating that the catalytic triad of R147A was not affected by the mutation.29 In contrast, the 2-step FX activation (chromogenic assay) showed that R147A and K192A demonstrated greatly reduced FX activation relative to WT, whereas the combined R147A/K192A showed a marked decrease in FX activation. The thrombin generation assay demonstrated comparable peak thrombin and ETP values, whereas significant prolonged lag time and time-to-peak values were noticed in FVII variants compared with WT. This observation could potentially be due to the “inefficiency” of FVII variants to bind to FX during the initiation phase of coagulation, as demonstrated by prolonged lag time and time to peak. The lag times became progressively prolonged with decreasing extrinsic factor concentrations.30 In addition, this effect was more remarkable in the presence of low concentration of TF (1 pM). In our thrombin generation experiment, when FVII variants were able to adequately generate FXa and cofactors, the intrinsic tenase complex was able to function properly and yielded normal thrombin generation as shown by similar peak thrombin and ETP compared with WT. Thrombin generation curves were also measured at 0.5- and 2-nM FVII variants in the presence of 0.4-pM TF and 4-μM phospholipids. All thrombin generation parameters also demonstrated the same pattern (data not shown). Our results were consistent with a previous study showing that peak thrombin and ETP were similar, but lag times were prolonged in FVII-deficient plasma spiked with different amounts of normal plasma at low (1 pM) and high (5 pM) concentrations of TF.30 In addition, the clotting time was markedly prolonged with R147A and K192A compared with WT, and no FVII coagulant activity was measurable in R147A/K192A. The reduced ability of FVII variants to activate FX as demonstrated by the chromogenic assay was consistently reproduced in the clotting assay. At this point, we summarized that the reduction of enzymatic and coagulant activities of FVII variants caused by TFPI-binding exosite mutations was not likely the result of impaired protease function.
TFPI is a strong competitive inhibitor of FVII.31 This implies that the amino acids within the substrate-binding pocket of FVII are important for both FX and TFPI binding. To study the importance of TFPI-binding exosites of FVII, the amino acids in the substrate-binding pocket of FVIIa involved in the binding with the Kunitz-1 domain of TFPI were analyzed through a structural analysis and sequence comparison using Pymol (Figure 6A).31 As depicted, D11 on the TFPI formed a salt bridge with R147 and K192 on FVIIa. The amino acid sequences of the Kunitz-1 domain of TFPI and the cleavage site of FX, where FVIIa binds and cleaves, were compared, and it was illustrated that FX possesses asparagine at the equivalent site to D11 on TFPI (Figure 6B). Consequently, FX cannot form a salt bridge with FVIIa, and the additional affinity between TFPI and FVII results in a strong competitive inhibitor.31 In fact, the Km values of FVII for FX were in the micromolar range,32,33 and the Ki values of TFPI for FVII were in the nanomolar range.34,35
Molecular model of human FVIIa crystal structure with the Kunitz-1 domain of TFPI. (A) Molecular model of FVIIa crystal structure with the Kunitz-1 domain of TFPI created in Pymol. The hydrogen bond network between residues within the substrate-binding pocket of FVIIa and the Kunitz-1 domain of TFPI adapted from PDB ID: 1FAK.31 R147 and K192 were illustrated as cyan stick. The Kunitz-1 domain of TFPI was shown as yellow stick. The catalytic triads in the active site were shown in magenta. Other residues within the substrate-binding pocket were shown in green. Hydrogen bonds were shown as black dotted lines. (B) The amino acid sequences at the cleavage sites of FX, binding with FVIIa, were compared with the Kunitz-1 domain of TFPI. The amino acids of FX and TFPI bound to R147 and K192 of FVIIa were highlighted in gray.
Molecular model of human FVIIa crystal structure with the Kunitz-1 domain of TFPI. (A) Molecular model of FVIIa crystal structure with the Kunitz-1 domain of TFPI created in Pymol. The hydrogen bond network between residues within the substrate-binding pocket of FVIIa and the Kunitz-1 domain of TFPI adapted from PDB ID: 1FAK.31 R147 and K192 were illustrated as cyan stick. The Kunitz-1 domain of TFPI was shown as yellow stick. The catalytic triads in the active site were shown in magenta. Other residues within the substrate-binding pocket were shown in green. Hydrogen bonds were shown as black dotted lines. (B) The amino acid sequences at the cleavage sites of FX, binding with FVIIa, were compared with the Kunitz-1 domain of TFPI. The amino acids of FX and TFPI bound to R147 and K192 of FVIIa were highlighted in gray.
R147 and K192 of FVII could form hydrogen bonds with asparagine (Figure 6B31) of FX before FX activation. R147 is essential in the extended recognition of macromolecular substrate FX to activate FX to FXa.29 It was reported that the R147A mutant decreased FX activation with a four- to fivefold and >10-fold reduction in the kcat in the presence and absence of phospholipid, respectively,29 and the R147A mutant was less inhibited by TFPI.36 FVII is important for TF-triggered thrombin generation because it is the protein that binds with the TF, and the TF–FVIIa complex activates FX to FXa in the initiation phase of the coagulation cascade.37 Therefore, mutation on these amino acids could weaken FX binding, leading to impaired FX activation, hence lowering FXa synthesis. The lower FXa being generated (Figure 5) led to a decreased thrombin generation and ultimately decreased fibrin formation.
In conclusion, our study deciphered the underlying mechanisms of bleeding phenotypes caused by the alteration of TFPI-binding exosites, R147 and K192, of FVII. The mutations resulted in a decrease in FVII secretion due to ER protein retention and ER stress. In addition, the FVII variants cannot properly bind to FX, resulting in reduction of FX activation. These data provided novel knowledge on the pathogenic mechanisms of FVII variants causing FVII deficiency. Moreover, our results may provide a fundamental concept regarding the ER stress pathway as therapeutic agents especially for patients with FVII deficiency and also for those with other congenital vitamin K–dependent coagulation factor deficiencies.
Acknowledgments
The authors would like to thank Pantep Angchaisuksiri for great support in reagents and equipment for thrombin generation assay and Sarai Pongjantarasatian for great help in cell culture techniques.
This work was financially supported by the Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (grant number RGNS 63-163), the Development and Promotion of Science and Technology Talents Project (DPST), Mahidol University, and the Children Cancer Fund under the patronage of HRH Princess Soamsawali.
Authorship
Contribution: K.S. performed the experiments and wrote the manuscript; P.P. developed the molecular model of human FVIIa crystal structure with the Kunitz-1 domain of TFPI and interpreted data; P.V., N.S., and A.C. contributed to intellectual discussion and reviewed the manuscript; S.H. supervised and supported the overall project; and P.T. designed the overall research and discussed, interpreted, and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pansakorn Tanratana, Department of Pharmacology, Faculty of Science, Mahidol University, 272 Rama VI Road, Ratchathewi District, Bangkok, 10400, Thailand; e-mail: pansakorn.tan@mahidol.ac.th; and Suradej Hongeng, Department of Pediatrics, Faculty of Medicine Ramathibodi Hospital, Mahidol University, 270 Rama VI Road, Ratchathewi District, Bangkok, 10400, Thailand; e-mail: suradej.hon@mahidol.ac.th.
References
Author notes
Data are available on request from the corresponding authors, Pansakorn Tanratana (pansakorn.tan@mahidol.ac.th) and Suradej Hongeng (suradej.hon@mahidol.ac.th).