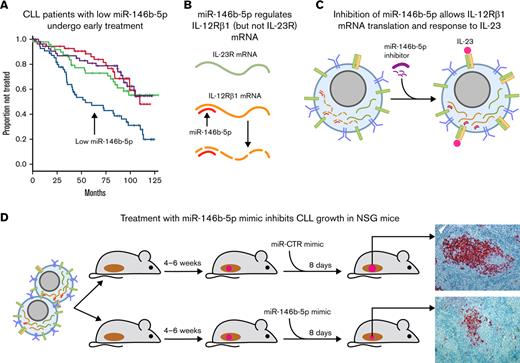
Key Points
Low concentrations of miR-146b-5p have an adverse prognostic impact in CLL patients.
miR-146b-5p controls IL-23 stimulation of CLL cells by negatively regulating the expression of the IL-12Rβ1 chain of the IL-23R complex.
Abstract
Chronic lymphocytic leukemia (CLL) cells express the interleukin-23 receptor (IL-23R) chain, but the expression of the complementary IL-12Rβ1 chain requires cell stimulation via surface CD40 molecules (and not via the B-cell receptor [BCR]). This stimulation induces the expression of a heterodimeric functional IL-23R complex and the secretion of IL-23, initiating an autocrine loop that drives leukemic cell expansion. Based on the observation in 224 untreated Binet stage A patients that the cases with the lowest miR-146b-5p concentrations had the shortest time to first treatment (TTFT), we hypothesized that miR-146b-5p could negatively regulate IL-12Rβ1 side chain expression and clonal expansion. Indeed, miR-146b-5p significantly bound to the 3′-UTR region of the IL-12Rβ1 mRNA in an in vitro luciferase assay. Downregulation of miR-146b-5p with specific miRNA inhibitors in vitro led to the upregulation of the IL-12Rβ1 side chain and expression of a functional IL-23R complex similar to that observed after stimulation of the CLL cell through the surface CD40 molecules. Expression of miR-146b-5p with miRNA mimics in vitro inhibited the expression of the IL-23R complex after stimulation with CD40L. Administration of a miR-146b-5p mimic to NSG mice, successfully engrafted with CLL cells, caused tumor shrinkage, with a reduction of leukemic nodules and of IL-12Rβ1–positive CLL cells in the spleen. Our findings indicate that IL-12Rβ1 expression, a crucial checkpoint for the functioning of the IL-23 and IL-23R complex loop, is under the control of miR-146b-5p, which may represent a potential target for therapy since it contributes to the CLL pathogenesis. This trial is registered at www.clinicaltrials.gov as NCT00917540.
Introduction
MicroRNAs (miRNAs) represent a family of noncoding RNAs that prevent the translation and promote the degradation of specific mRNAs by binding to their 3′-UTR.1,2 Several miRNAs have been implicated in the pathogenesis of chronic lymphocytic leukemia (CLL),3-5 a disease characterized by the accumulation of monoclonal CD5+CD19+ B cells in lymphoid organs and blood.6-9 In patients with 13q deletions (del[13q]), the most common cytogenetic lesion of CLL,10,11 the genes encoding the miR-15a/miR-16-1 cluster are targeted by the deletion.3,12-15 The downregulation of these regulatory miRNAs can lead to an increased expression of antiapoptotic molecules, which facilitate clonal expansion, inducing further transforming events.12-16 MiRNA expression profile studies have disclosed correlations between certain miRNA signatures and cytogenetic features and/or IGHV gene mutational status,17-19 which represent recognized prognostic markers of CLL. Finally, certain miRNA signatures are associated with disease progression and outcome4,17,20-22 or with the onset of a Richter transformation,23-25 a deadly condition characterized by the development of an aggressive lymphoma in CLL patients.7,26
Previously, we reported an inverse correlation between miR-146b-5p concentrations and progression-free survival in a cohort of >200 newly diagnosed Binet stage A patients; cases with the most aggressive clinical course had the lowest miR-146b-5p concentrations.17 The same inverse correlation was not observed with miR-146a-5p, a paralog of miR-146b-5p, in the same patient cohort. Although not validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR), these differences were substantial and somewhat surprising, given that the 2 miRNAs share many predicted target genes and the same seed sequence.27 However, the 2 miRNAs are encoded by genes located on different chromosomes (chromosome 5 and 10 for miR-146a-5p and miR-146b-5p, respectively), which may create differences in the posttranscriptional processing associated with the 2 other nucleotides encoded at the 3′ end.27 Another surprising difference was that the CLL cases with the lowest miR-146b-5p concentrations were also IGHV-unmutated (UM), while this correlation was not observed in the case of miR-146a-5p.
Both miR-146a-5p and miR-146b-5p control the proliferation of a variety of cells, particularly because they regulate NF-kB (nuclear factor kappa B) activation, a key transcription factor involved in cell proliferation.28,29 Both miR-146a-5p and miR-146b-5p exert a negative regulatory control on the expression of TNFR6 (tumor necrosis factor receptor-6) and IRAK1 (interleukin-1 receptor-associated kinase 1), 2 adaptor molecules that transduce signals delivered via several membrane receptors, such as those of the TNFR and the Toll-like receptor/IL1R superfamilies27,30-32 culminating in NF-kB activation. This function accounts in part for the spontaneous onset of cancers in mice with deletions of miR-146a-5p33,34 and the inverse correlation reported in human cancers between tumor aggressiveness and miR-146b-5p concentrations.35-40 Inflammatory and autoimmune phenomena observed in mice with deletions of these miRNAs may also be explained by an absent NF-kB regulation.34,41-45 However, the observation that miR-146b-5p is more effective than miR-146a-5p in determining CLL clinical course17 suggests that miR-146b-5p is implicated in additional mechanisms supporting CLL clonal expansion that are different from TRAF6 (tumor necrosis factor receptor-associated factor 6) and IRAK1 control.
Considerable evidence indicates that CLL clonal expansion is promoted by interactions with cells and cytokines from the microenvironment.46,47 Moreover, both miR-146a-5p and miR-146b-5p can regulate the release of and the response to cytokines.41,48,49 Based on these considerations, we hypothesized that miR-146b-5p was involved in the regulation of the interactions between CLL cells and the microenvironment. We focused on IL-23,50 a cytokine of the IL-12 cytokine family, released primarily by dendritic cells, which is capable of driving T helper (Th) cell differentiation toward the Th17 cell subset.51 In a previous study, we found that IL-23 is instrumental in promoting CLL cell proliferation and clonal expansion.50 Normally, circulating CLL cells express variable concentrations of the IL-23R chain, 1 of the 2 chains forming the heterodimeric IL-23R complex, but are incapable of responding to IL-23 because of the absence of its complementary chain, IL-12Rβ1. Upon appropriate activation signals in vitro, such as the interaction with activated T cells or other CD40L-expressing cells, but not via direct stimulation of the B-cell receptor (BCR), CLL cells express the IL-12Rβ1 chain and begin to secrete IL-23.50 This initiates an autocrine/paracrine loop (which we have named the IL-23/IL-23R complex loop), whereby CLL cells respond to the IL-23 that they produce. This event promotes leukemic cell proliferation50 and appears to be very relevant for CLL cell growth/expansion since most leukemic cells in the proliferating centers of lymphoid tissues, infiltrated by CLL cells, produce IL-23 and express a complete IL-23R complex.50 Moreover, in vivo treatment with antibodies to IL-23p19 (1 of the 2 chains forming the IL-23 molecules) eradicates CLL clones in xenografted mice.50 Because the expression of the IL-12Rβ1 chain by CLL may represent a key checkpoint for the initiation of the loop, we hypothesized that miR-146b-5p was involved in regulating the expression of this chain. Indeed, the present findings support our hypothesis and show that miR-146b-5p can be a key regulator in controlling CLL cell clonal expansion.
Methods
Patients and CLL cell preparations
The patients investigated were part of the O-CLL1 study (clinicaltrials.gov identifier NCT00917540), an observational cohort of patients with untreated Binet A CLL collected from several Italian institutions enrolled within 12 months from diagnosis.17,52 Supplemental Table 1 in the data supplement summarizes the clinical features of the patients investigated.52-54 In total, samples from 224 CLL cases were studied for expression profiles and single miR expression17,52; the data are deposited at the NCBI (National Center for Biotechnology Information) GEO (Gene Expression Omnibus) repository (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE40533. For CLL cases not included in the miRNome study, we measured miR-146b-5p concentrations by quantitative real-time PCR (RT-qPCR). Also, for these cases, miR-146b-5p expression was significantly correlated with immunoglobulin heavy chain variable region (IGVH) gene mutational status (see supplemental Methods, supplemental Table 2, and supplemental Figure 1). Peripheral blood mononuclear cells from patients with CLL were isolated by Ficoll-Hypaque (Seromed, Biochrom) density gradient centrifugation, and CD19-positive CLL cells were enriched by negative selection as previously reported50 (see supplemental Methods). Written informed consent was obtained from all patients in accordance with the declaration of Helsinki. The ethics committees from each participating center (listed in the acknowledgments) approved this study.
Viable cell counts of CLL samples were conducted before each experiment performed in vitro and in vivo by trypan blue staining and automatic cell counter (Countess, Invitrogen). Values >80% of live cells were considered suitable for the subsequent experimental procedures.
Cell transfection
MirVana miRNA mimics or inhibitors (Ambion Inc, Thermo Fisher Scientific; Grand Island, NY) were delivered to CLL cells using the Neon Transfection System (Invitrogen, Thermo Fisher Scientific) as described15 or by the Nucleofector-4D Transfection System (Amaxa), (supplemental Methods). The following miRNA mimics and inhibitors were used: hsa-miR-146b-5p (Assay ID: MC25960; MH25960), hsa-miR-146a-5p (Assay ID: MC10722), miRNA mimic, Negative Control#1 (Cat. no. 4464058), miRNA inhibitor, Negative Control #1 (Cat. no. 4464076). The transfection efficiency was verified by RT-qPCR (see supplemental Methods).
Detection of the IL-23R complex
Cell surface IL-12Rβ1 and IL-23R chains were detected by flow cytometry.50 IL-12Rβ1 expression also was analyzed by Western blotting with mouse anti–IL-12Rβ1 monoclonal antibody (mAb) (C-20, sc-658, Santa Cruz Biotechnology, Inc.) and an anti-GAPDH mAb (AM4300, Ambion Inc, Thermo Fisher Scientific) as a loading control. qRT-PCR assessed the IL-12Rβ1 side chain mRNA (see supplemental Methods).
Luciferase reporter assays
MiRNA target reporter vectors were purchased from Origene (IL-12Rβ1, Accession No. NM_153701, transcript variant 2, Cod. SC208722) and Switchgear (IL-23R, Cat. S806498, Accession No. NM_144701). IL-12Rβ1-MUT reporter vector, obtained by deletion of miR-146b-5p seed target site sequence (GTTCTCA [nt328-nt334]), was custom produced by Origene (Figure 2C). 3′ UTR assays are described in the supplemental Methods. HEK293 cells were used for transfection and the luciferase reporter assays. Preliminary tests showed that CLL-related cell lines (MEC-1 and OSU cell lines) were not suitable for testing because of the poor yield of the transfection step.
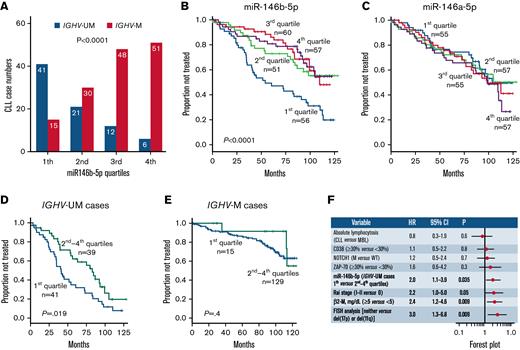
Capacity of miR-146b-5p concentrations of predicting TTFT in CLL. (A) Association between miR-146b-5p concentrations and IGHV mutational status in 224 CLL cases. The numbers of CLL cases (subdivided into different quartiles according to miR-146b-5p concentrations) are indicated in each bar. (B) Kaplan-Meier curves of the 6-year TTFT probability of cases stratified according to miR-146b-5p expression. Cases with the lowest miR-146b-5p (first quartile) expression had a 47% probability of prolonged TTFT compared with that of 73%, 83%, and 79% of cases within the second, third, and fourth quartile, respectively. Statistical significance of associations between individual variables and survival was calculated using the log-rank test. (C) Kaplan-Meier curves showing TTFT probability of cases stratified in different quartiles defined by miR-146a-5p expression. Statistical significance of associations between individual variables and survival was calculated using the log-rank test. (D) Kaplan-Meier curves comparing TTFT of miR-146b-5p–low (first quartile) or miR-146b-5p higher expression (second to fourth quartiles) in IGHV-UM cases and in (E) IGHV-M cases. Statistical significance of associations between individual variables and survival was calculated using the log-rank test. (F) Cox multivariate analysis of IGVH-UM CLL cases (n = 80) showing that low miR-146b-5p expression (first quartile) maintains an independent prognostic impact in the presence of other prognostic indicators (P = .035).
Capacity of miR-146b-5p concentrations of predicting TTFT in CLL. (A) Association between miR-146b-5p concentrations and IGHV mutational status in 224 CLL cases. The numbers of CLL cases (subdivided into different quartiles according to miR-146b-5p concentrations) are indicated in each bar. (B) Kaplan-Meier curves of the 6-year TTFT probability of cases stratified according to miR-146b-5p expression. Cases with the lowest miR-146b-5p (first quartile) expression had a 47% probability of prolonged TTFT compared with that of 73%, 83%, and 79% of cases within the second, third, and fourth quartile, respectively. Statistical significance of associations between individual variables and survival was calculated using the log-rank test. (C) Kaplan-Meier curves showing TTFT probability of cases stratified in different quartiles defined by miR-146a-5p expression. Statistical significance of associations between individual variables and survival was calculated using the log-rank test. (D) Kaplan-Meier curves comparing TTFT of miR-146b-5p–low (first quartile) or miR-146b-5p higher expression (second to fourth quartiles) in IGHV-UM cases and in (E) IGHV-M cases. Statistical significance of associations between individual variables and survival was calculated using the log-rank test. (F) Cox multivariate analysis of IGVH-UM CLL cases (n = 80) showing that low miR-146b-5p expression (first quartile) maintains an independent prognostic impact in the presence of other prognostic indicators (P = .035).
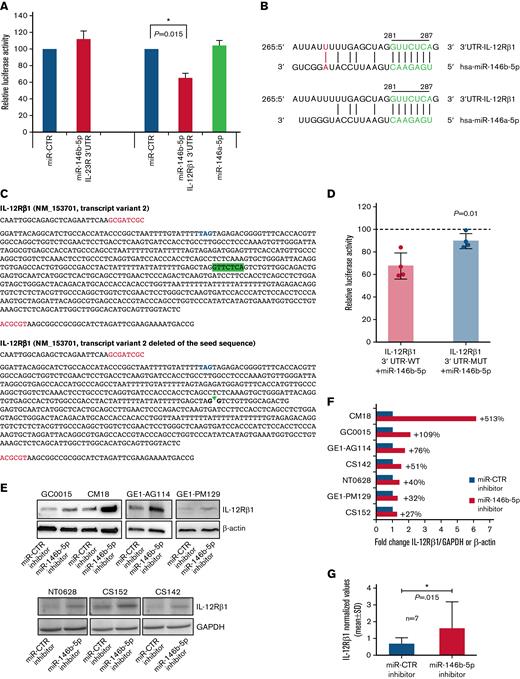
Potential regulatory function of miR-146b-5p on the expression of IL-12Rβ1. (A) The inhibitory effect of a given miRNA on a target sequence (3′UTR) expression was measured in cultured HEK293 cells. These cells were transfected with either the IL-23R 3′UTR or the 3' UTRI L-12Rβ1 together with miR mimics and miR control as indicated. Both firefly luciferase and Renilla luciferase activities were measured after a 48-hour culture. Data shown are relative to the reporter vector transfected with the miR CTR mimic and represent the mean of 5 and 7 experiments, respectively, carried out in triplicate. ∗P < .05. (B) Sequence alignment of miR-146b-5p and miR-146a-5p. The seed region on miRNAs and the potential target sequence on mRNA (site type 7mer-A1, Target scan release 7.2) are indicated in green. A further base pairing of miR-146b-5p with the 3′UTR IL-12Rβ1 is indicated in red. The position coordinates are indicated for the IL-12Rβ1 transcript isoform 3′ UTR: ENSG0000096996.11: ENST00000322153.7. (C) The insert sequence of 3′ UTR clone NM_153701 (Origene, cod SC208722) (IL-12Rβ1 3' UTR-WT) and the same insert deleted of the sequence GTTCTCA complementary to the seed sequence (nt328-nt334, Origene) (IL-12Rβ1 3′ UTR-MUT). Blue, stop codon; red, cloning site; highlighted in green, the seed sequence. (D) HEK 293 cells were transfected with either the IL-12Rβ1 3′UTR-WT or with IL-12Rβ1 3′UTR-MUT with miR mimics and miR control. Both firefly luciferase and Renilla luciferase activities were measured after a 48-hour culture. Data shown are relative to the reporter vector transfected with the miR CTR mimic (reference line at 100%) and represent the mean of 4 experiments carried out in triplicate. P value is statistically significant (P < .05, t test). (E) Western blotting analysis of IL-12Rβ1 and GAPDH or β-Actin expression in purified CLL cells from 7 different cases transfected with miR-146b-5p inhibitor or miR-CTR inhibitor and cultured for 48 hours. (F) Fold change values of the IL-12Rβ1 signal normalized to that of GAPDH or β-Actin of cells transfected with miR-146b-5p inhibitor/IL-12Rβ1 normalized signal of cells transfected with miR-CTR inhibitor. Percentage (%) changes in the relative abundance of IL-12Rβ1 protein are also indicated for each CLL sample and calculated as (fold change − 1) × 100. A positive percentage indicates increased abundance relative to the control. (G) Summary of the results of the experiments in (F). Protein bands from immunoblotting were analyzed using ImageJ Analysis Software or by Alliance LD, UVITEC. Data are presented as IL-12Rβ1/GAPDH or IL-12Rβ1/β-Actin (mean ± SD). The P value of the difference between CLL cells treated with miR-146b-5p vs miR-CTR inhibitors is indicated (Wilcoxon test). ∗P < .05.
Potential regulatory function of miR-146b-5p on the expression of IL-12Rβ1. (A) The inhibitory effect of a given miRNA on a target sequence (3′UTR) expression was measured in cultured HEK293 cells. These cells were transfected with either the IL-23R 3′UTR or the 3' UTRI L-12Rβ1 together with miR mimics and miR control as indicated. Both firefly luciferase and Renilla luciferase activities were measured after a 48-hour culture. Data shown are relative to the reporter vector transfected with the miR CTR mimic and represent the mean of 5 and 7 experiments, respectively, carried out in triplicate. ∗P < .05. (B) Sequence alignment of miR-146b-5p and miR-146a-5p. The seed region on miRNAs and the potential target sequence on mRNA (site type 7mer-A1, Target scan release 7.2) are indicated in green. A further base pairing of miR-146b-5p with the 3′UTR IL-12Rβ1 is indicated in red. The position coordinates are indicated for the IL-12Rβ1 transcript isoform 3′ UTR: ENSG0000096996.11: ENST00000322153.7. (C) The insert sequence of 3′ UTR clone NM_153701 (Origene, cod SC208722) (IL-12Rβ1 3' UTR-WT) and the same insert deleted of the sequence GTTCTCA complementary to the seed sequence (nt328-nt334, Origene) (IL-12Rβ1 3′ UTR-MUT). Blue, stop codon; red, cloning site; highlighted in green, the seed sequence. (D) HEK 293 cells were transfected with either the IL-12Rβ1 3′UTR-WT or with IL-12Rβ1 3′UTR-MUT with miR mimics and miR control. Both firefly luciferase and Renilla luciferase activities were measured after a 48-hour culture. Data shown are relative to the reporter vector transfected with the miR CTR mimic (reference line at 100%) and represent the mean of 4 experiments carried out in triplicate. P value is statistically significant (P < .05, t test). (E) Western blotting analysis of IL-12Rβ1 and GAPDH or β-Actin expression in purified CLL cells from 7 different cases transfected with miR-146b-5p inhibitor or miR-CTR inhibitor and cultured for 48 hours. (F) Fold change values of the IL-12Rβ1 signal normalized to that of GAPDH or β-Actin of cells transfected with miR-146b-5p inhibitor/IL-12Rβ1 normalized signal of cells transfected with miR-CTR inhibitor. Percentage (%) changes in the relative abundance of IL-12Rβ1 protein are also indicated for each CLL sample and calculated as (fold change − 1) × 100. A positive percentage indicates increased abundance relative to the control. (G) Summary of the results of the experiments in (F). Protein bands from immunoblotting were analyzed using ImageJ Analysis Software or by Alliance LD, UVITEC. Data are presented as IL-12Rβ1/GAPDH or IL-12Rβ1/β-Actin (mean ± SD). The P value of the difference between CLL cells treated with miR-146b-5p vs miR-CTR inhibitors is indicated (Wilcoxon test). ∗P < .05.
CLL cell cultures
After transfection with the appropriate miRNA, CLL cells were cultured in RPMI 1640 medium with γ-irradiated50 cells from a stable CD40L-expressing NIH-3T3 (CD40L-TC) murine fibroblast cell line or with the NIH-3T3 cells transfected with the pIRES empty vector (Mock) (1 NIH-3T3 cell: 100 CLL cells) at a concentration of 2 × 106 cells per mL at 37°C in an atmosphere containing 5% CO2.
IL-23 detection
IL-23 cytokine production was measured in cell culture supernatants using the Human Cytokine/Chemokine Panel II and Luminex MAGPIX System (Merck Millipore).
Xenogeneic mouse transplantation
These procedures were described previously,15,50,55 and additional details are provided in the supplemental Methods. All animal experiments were performed according to the current national and international regulations and were approved by the Licensing and Animal Welfare Body of the IRCCS-Ospedale Policlinico San Martino, Genoa, Italy.
Statistical analysis
The statistical package SPSS for Windows (release 13.0, 2004 software, SPSS UK; Surrey, United Kingdom) was used for all analyses. Statistical comparisons were performed using 2-way tables for the Fisher’s exact test and multiway tables for the Pearson’s χ2 test. Statistical comparisons between related samples were carried out by Wilcoxon or Mann-Whitney U tests. Time-to-first treatment (TTFT) analyses were performed using the Kaplan-Meier method. Statistical significance of associations between individual variables and survival was calculated using the log-rank test. The prognostic impact for the outcome variable was investigated by univariate and multiple Cox regression analysis. Data are expressed as hazard ratio (HR) and 95% confidence intervals (CIs). A value of P < .05 was considered significant for all statistical calculations. Values are given as mean ± SD.
Results
Predictive power of miR-146b-5p expression by CLL cells
First, we confirmed that miR-146b-5p concentrations maintained their prognostic power using a large CLL cohort described previously (O-CLL1 protocol).52 This comprised 224 Binet stage A patients, 48 of whom met the current diagnostic criteria of clinical monoclonal B-lymphocytosis.52,56 As shown in Figure 1A, miR-146b-5p was less expressed in CLL cases with IGHV-UM genes than in those with mutated IGHV (IGHV-M). The majority (41 of 56 [73%]) of cases with the lowest miR-146b-5p concentrations (first quartile) were IGHV-UM (Figure 1A). The median follow-up time in the cohort investigated was 83 months (range, 1-129), and 94 patients had progressed and required therapy at the time of the study censoring. Cases within the quartile with the lowest miR-146b-5p concentrations (first quartile) also had the shortest TTFT (Figure 1B). MiR-146a-5p failed to identify patients with a shorter TTFT (Figure 1C), a finding consistent with the observation that the concentrations of miR-146a-5p were similar in IGHV-M (n = 144, mean ± SD = 49 ± 95) and IGHV-UM cases (n = 80, mean ± SD = 45 ± 47) (supplemental Figure 2A-B). Furthermore, no correlation was observed between the expression of miR-146b-5p and miR-146a-5p, although the differences in expression between quartiles were similar for both miRNAs (supplemental Figure 2C-D).
In a Cox multivariate model, together with other prognostic markers (IGHV-UM, CD38-positive, ZAP-70–positive, mutated NOTCH1 gene, RAI stage, FISH del(17p) or del(11q), β2-microglobulin (β2-M) values ≥5 mg/dL, and patients with a peripheral B-lymphocytosis of ≥5000/mm3), low miR-146b-5p expression failed to predict TTFT (supplemental Tables 3 and 4, Model 1). However, following the stratification of cases according to the IGHV mutational status, IGHV-UM cases with the lowest miR-146b-5p concentrations (first quartile) had TTFT curves that were significantly different from those of cases in the remaining quartiles. These differences were not observed in IGHV-M cases (Figure 1D-E). The analysis of the quartiles calculated within each IGHV-M and IGHV-UM group demonstrates the consistent survival association only within the IGHV-UM group (supplemental Figure 3). Cox multivariate analysis, with the variables used above, demonstrated a significant independent association between low concentrations of miR-146b-5p and clinical outcome (HR, 2.0; 95% CI, 1.1-3.9; P = .035) (Figure 1F) in IGHV-UM cases. Observations in 21 pairs of CLL cell samples taken from the same patients at disease onset and progression showed no changes in miR-146b-5p concentrations at disease progression (supplemental Figure 4).
Analysis of mutations, CNA, and methylation status of the miR-146b-5p locus
To investigate possible mutations and copy number alterations (CNAs) on miR-146b and its putative promoter/enhancer regions possibly responsible for the occurrence of lower concentrations of miR-146b-5p in a subset of patients with CLL, we analyzed a dataset of 551 patients with CLL (CLLE-ES)57 by ICGC (International Cancer Genome Consortium) Data Portal (release_28),58 that collects sequencing data from different repositories, including the European genome–phenome archive. No patients with CLL presented somatic mutations in miR-146b genomic region (chr10:104196269-104196341) or putative promoter/enhancer regions predicted by GeneHancer59 (supplemental Table 5).
CNA analysis performed on the same patients with CLL dataset showed the existence of a loss of the genomic region, including miR-146b, in 7 of 551 (1.3%) patients. CNA coordinates and patient characteristics are reported in supplemental Table 6. Similar results were obtained by Leeksma and colleagues,60 who retrospectively analyzed 2293 arrays for CNA assessment from 13 diagnostic laboratories according to established standards and found 10q losses in 25 of 2293 patients (approximately 1%). About half of these (13 of 2293 [0.6%]) showed 10q deletion encompassing miR-146b at the 10q24.32 locus. Therefore CNA at the miR-146b locus could not account for our observations.
We then investigated the possibility that miR-146b-5p expression in CLL could be epigenetically regulated. Methylation status of miR-146b locus was explored in CLL cases, and normal B-cell samples by whole-genome bisulfite sequencing reported in the BluePrint Data Analysis portal (http://dcc.blueprint-epigenome.eu),61 considering a region spanning 500 bp upstream and downstream the miR-146b locus, respectively (GRCh37.p13 chr10:102436500-102436609, EnsEMBL version: 79).
In CLL samples, mean concentrations of hypomethylation or emimethylation were detected in the region upstream or in the 150 bp immediately downstream miR-146b locus, whereas hypermethylation was found in the region encompassing miR-146b and in more downstream regions. In naïve and memory B cells from peripheral blood, respectively, a global pattern of hypomethylation was evidenced in the upstream regions, whereas hypermethylation was observed downstream to miR-146b locus (supplemental Figure 5). Therefore, a wider range and higher methylation concentrations than normal were observed in the upstream region of the miR-146b locus in CLL samples.
In addition, the DNA methylation of the miR-146b-5p gene suggests that DNA methylation is directly involved in the regulation of its biogenesis.62 This regulation could be dependent on activating stimuli received by neoplastic cells in the lymphoid organs.
Regulation of IL-23R complex expression by miR146b-5p in CLL cells
We next investigated whether miR-146b-5p could regulate the expression of IL-23R and/or IL-12Rβ1 chains. CLL clones can be subdivided into those with a low (IL-23R–low) or a high level of IL-23R (IL-23R–high) expression, respectively, when stratified according to a cutoff of IL-23R chain-positive cells lower or greater than 23%.50 We performed a correlation analysis to ascertain whether miR-146b-5p was lower in cases with higher IL-23R expression in a group of 93 CLL patients (40 cases IL-23R–low and 53 cases IL-23R–high). Although a significant anticorrelation in expression was detected (RHO−0.291; P = .005) (supplemental Figure 6), in vitro luciferase reporter assay failed to demonstrate a significant binding of miR-146b-5p to the IL-23R 3′UTR mRNA (Figure 2A).
We used a recently developed web tool named miRabel (http://bioinfo.univ-rouen.fr/mirabel/)63 to investigate the potential binding of miR-146b-5p to the 3′UTR of the IL-12Rβ1 chain mRNA (for details, see supplemental Methods). This approach predicted a substantial binding capacity (score 0.3572489917218290), which was confirmed experimentally in luciferase reporter assays (Figure 2A), showing an average reduction of the luciferase activity of 35 ± 10.3% (mean ± SD). In contrast, miR-146b-5p did not efficiently bind the IL-12Rβ1 3′UTR–MUT with an average reduction of the luciferase activity of 10 ± 6% vs 33 ± 11% of the 3′UTR WT (mean ± SD; P = .01) (Figure 2C-D) confirming the specificity of the interaction between the miR-146b-5p seed sequence and the complementary sequence on the IL-12Rβ1 chain mRNA. The possible binding of miR-146a-5p to the 3′UTR of the IL-12Rβ1 chain mRNA, predicted by the same algorithms, was not confirmed experimentally (Figure 2). To further confirm the miRNA-mediated regulation IL-12Rβ1 side chain, primary CLL cells were transiently transfected with a specific miRNA inhibitor targeting miR-146b-5p or with a miR-control inhibitor (a random sequence molecule with no identifiable effects on known miRNA functions) and cultured for 48 hours. A consistent upregulation of the IL-12Rβ1 side-chain protein by knocking down miR-146b-5p expression was found by Western blot (Figure 2E-G).
Upregulation of the IL-12Rβ1 chain and expression of a functional IL-23R complex in CLL cells following downregulation of miR-146b-5p
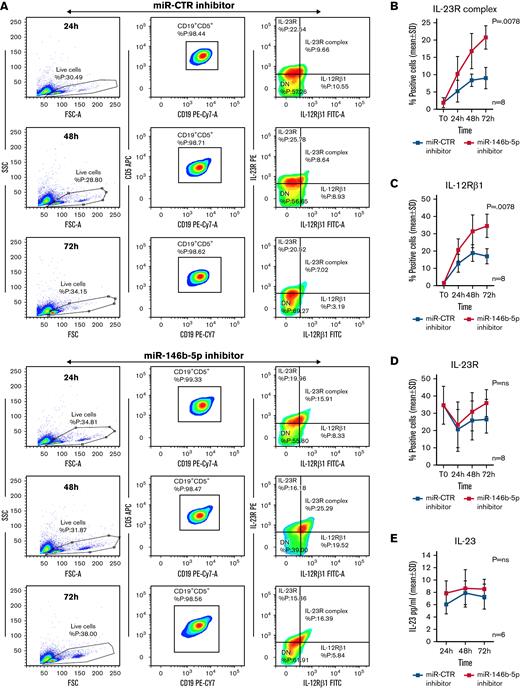
The above target validation experiments prompted tests aimed at verifying whether miR-146b-5p inhibition could induce the expression of the Il-12Rβ1 side chain and a functional IL-23R complex on the surface of CLL clones already expressing an IL-23R side chain. Purified CLL cells from 8 IL-23R–high cases (35 ± 11% [mean ± SD] positive cells) (GE1-AG114, GE1-DM210, GC0015, SV1-SA, SR1-ME1077, MG0482, VF0384, and CM18) were transiently transfected with a miR-146b-5p inhibitor or with a miR-CTR inhibitor, cultured for different times, and tested for IL-12Rβ1 and IL-23R side chains expression. Cells transfected with miR-146b-5p inhibitor had a significantly increased expression (P = .0078) of IL-23R complex (average increase value of 57 ± 12% positive cells at 72 hours in culture [mean ± SD]) compared with the control cells (Figure 3A-B). The increased IL-23R complex expression was associated with an upregulation of the IL-12Rβ1 side chain (average increase of 51 ± 9% positive cells at 72 hours [mean ± SD]) that was significantly different (P = .0078) from the control samples; in contrast, the expression of the IL-23R side chain remained virtually unchanged (Figure 3C-D). The cells positive for the chains of the IL-23R complex were identified within the gated populations of viable cells (Figure 3A and supplemental Figure 7). IL-23 released by CLL cells in the culture supernatants was also measured. There were no differences in the IL-23 produced by the miR-146b-5p inhibitor transfected and control cells (Figure 3E).
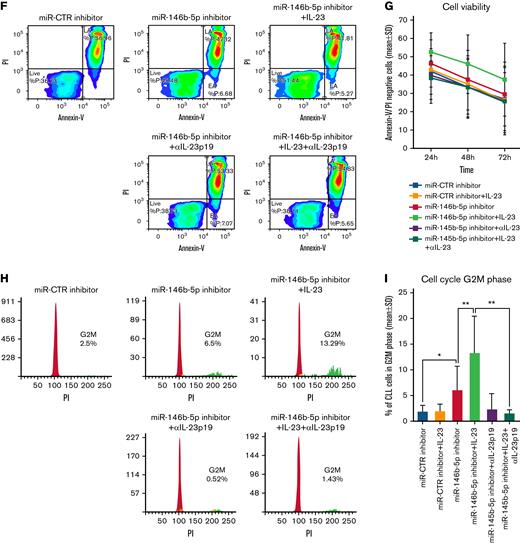
Expression of the IL-12Rβ1 chain and a functional IL-23R complex following downregulation of miR-146b-5p. Purified CLL cells were transfected with miR-146b-5p inhibitor or miR CTR inhibitor, cultured for different times and analyzed for the IL-23R complex expression and for IL-23 production. (A) Time course analysis of the IL-23R complex expression after transfection with miR CTR inhibitor or miR-146b-5p inhibitor in a representative CLL case (GE1-AG114). Cells expressing the IL-23R complex were measured by flow cytometry, determining the simultaneous expression of both the IL-23R and IL-12Rβ1 chains. Double-positive cells were considered as IL-23R complex-positive. Only viable cells were gated, and of these were mainly CD19+CD5+ since cells with this phenotype that were purified before transfection (see also supplemental Figure 7). (B-D)Summary of time course expression of IL23R complex, (C) IL-12Rβ1, and (D) IL-23R side chain determined in cells from 8 CLL cases by flow cytometry before (T0) and after treatment as in (A). Data are expressed as a percentage of positive cells (mean ± SD). (E) IL-23 production in cell supernatants from 6 CLL cases treated as in (A). (F) Representative experiment on cells from GE1-AG114 CLL case to show the presence of a functional IL-23R complex. Purified CLL cells were transfected with the indicated miR inhibitors and cultured for 24 to 72 hours in the presence or absence of IL-23 (100 ng/mL), with/without IL-23-neutralizing mAbs (αIL-23p19). Viable cells (annexin-V/PI-negative cells) were determined after a 72-hour culture. (G) Summary of time course experiments on cells from 8 CLL cases treated and analyzed as in (F). Data are plotted as percent of viable cells mean ± SD, and the P value indicates the differences between the different culture conditions. (H) Determination of cell cycle phases by flow cytometry in CLL cells (GE1-AG114) transfected with the indicated miR inhibitors and cultured for 48 hours in the presence or absence of IL-23 (100 ng/mL), with/without IL-23 neutralizing mAbs (αIL-23p19) in the indicated combinations. Flow logic software was employed for the analyses. Proliferating (G2M) cells are indicated in green. (I) Summary of experiments on cells from 8 different CLL cases performed and analyzed as in (I). The values were determined after 48 hours in culture. P values are indicated (Wilcoxon test). ∗P = .04 and ∗∗P = .0078.
Expression of the IL-12Rβ1 chain and a functional IL-23R complex following downregulation of miR-146b-5p. Purified CLL cells were transfected with miR-146b-5p inhibitor or miR CTR inhibitor, cultured for different times and analyzed for the IL-23R complex expression and for IL-23 production. (A) Time course analysis of the IL-23R complex expression after transfection with miR CTR inhibitor or miR-146b-5p inhibitor in a representative CLL case (GE1-AG114). Cells expressing the IL-23R complex were measured by flow cytometry, determining the simultaneous expression of both the IL-23R and IL-12Rβ1 chains. Double-positive cells were considered as IL-23R complex-positive. Only viable cells were gated, and of these were mainly CD19+CD5+ since cells with this phenotype that were purified before transfection (see also supplemental Figure 7). (B-D)Summary of time course expression of IL23R complex, (C) IL-12Rβ1, and (D) IL-23R side chain determined in cells from 8 CLL cases by flow cytometry before (T0) and after treatment as in (A). Data are expressed as a percentage of positive cells (mean ± SD). (E) IL-23 production in cell supernatants from 6 CLL cases treated as in (A). (F) Representative experiment on cells from GE1-AG114 CLL case to show the presence of a functional IL-23R complex. Purified CLL cells were transfected with the indicated miR inhibitors and cultured for 24 to 72 hours in the presence or absence of IL-23 (100 ng/mL), with/without IL-23-neutralizing mAbs (αIL-23p19). Viable cells (annexin-V/PI-negative cells) were determined after a 72-hour culture. (G) Summary of time course experiments on cells from 8 CLL cases treated and analyzed as in (F). Data are plotted as percent of viable cells mean ± SD, and the P value indicates the differences between the different culture conditions. (H) Determination of cell cycle phases by flow cytometry in CLL cells (GE1-AG114) transfected with the indicated miR inhibitors and cultured for 48 hours in the presence or absence of IL-23 (100 ng/mL), with/without IL-23 neutralizing mAbs (αIL-23p19) in the indicated combinations. Flow logic software was employed for the analyses. Proliferating (G2M) cells are indicated in green. (I) Summary of experiments on cells from 8 different CLL cases performed and analyzed as in (I). The values were determined after 48 hours in culture. P values are indicated (Wilcoxon test). ∗P = .04 and ∗∗P = .0078.
Next, we investigated the functional features of the IL-23R complex expressed by CLL cells. CLL cells purified from the same 8 patients were transiently transfected with the miR-146b-5p inhibitor or with the miR-CTR inhibitor and cultured in the presence or absence of recombinant IL-23 for different time points. Upon exposure to exogenous IL-23 in culture, significant increases (P = .0078) in cell viability (mean ± SD increase at 72 hours, 22.6 ± 10%) (Figure 3F-G) and of cycling cells (mean ± SD increase, 48.7 ± 27%) (Figure 3H-I) were observed in suspensions treated with the miR-146b-5p inhibitor; these effects were abrogated by the addition of a specific IL-23 mAb (αIL-23p19) to the culture supernatant (average inhibition at 72 hours, 37 ± 16%, for cell viability, and 84 ± 15% for cycling cells) (Figure 3F-I).
Downregulation of miR-146b-5p and expression of the IL-23R complex by CLL cells stimulated with CD40L
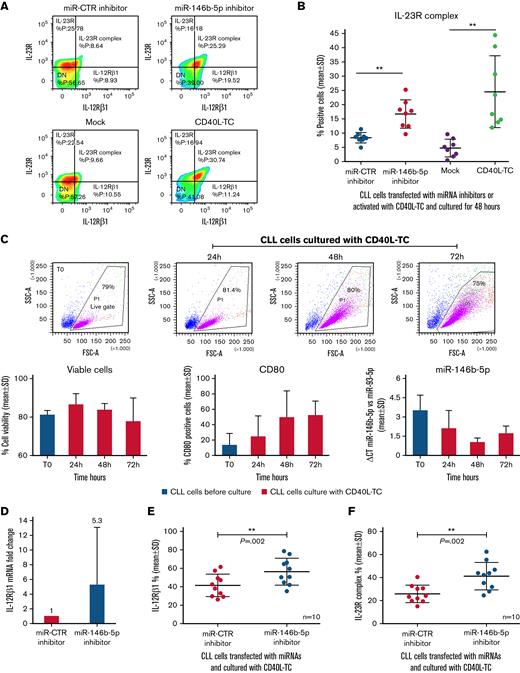
Since stimulation of CLL cells with CD40L in vitro induces the expression of a functional IL-23R complex,50 we investigated whether the same stimulation caused the downregulation of miR-146b-5p. Purified CLL cells from the 8 patients studied above were either transfected with the miR-146b-5p inhibitor or cultured with CD40L-TC. In both instances, the expression of the IL-12Rβ1 chain (and consequently of the IL-23R complex) was observed in amounts superior to those observed in the respective control cultures (average increase of 46 ± 19% for miR-146b-5p inhibitor treatment and of 77.3 ± 15.6% for CD40L-TC stimulation [mean ± SD], respectively) (Figure 4A-B). To investigate the concentrations of miR-146b-5p following CD40L-TC stimulation, purified CLL cells from 3 different cases with a variable baseline amount of miR-146b-5p were cultured with CD40L-TC and harvested at intervals. Viable cells were measured by flow cytometry by excluding annexin-V/PI-positive cells, whereas activated cells were identified as CD80+ cells (Figure 4C). Cell viability remained high throughout the culture, while there was a progressive acquisition of CD80 expression over time. Exposure to CD40L in vitro caused a substantial downregulation of miR-146b-5p as assessed by RT-qPCR (average inhibition at 48 hours, 70.1 ± 1.7% [mean±SD]) (Figure 4C). In contrast, stimulation of purified CLL cells by coculture with anti-μ and anti-δ Ig-chain–coated beads and IL-4 failed to significantly modify miR-146b-5p expression (supplemental Figure 8 and supplemental Methods). These data also are consistent with our previous findings on the incapacity of cell stimulation via BCR to induce the IL-23R complex expression.50
Downregulation of miR-146b-5p expression by CLL cells cocultured with CD40L-TC. (A) Comparison of IL-23R complex expression by cells from a representative CLL case (GE1-AG114) transfected with the indicated miR inhibitors or cocultured with CD40L-TC or mock control cells for 48 hours and analyzed by flow cytometry. (B) Summary of data on IL-23R complex expression (determined by flow cytometry) after a 48-hour exposure of purified CLL cells to miR-146b-5p inhibitor or CD40L-TC as in (A). CLL cells were from cases GE1-AG114, GE1-DM210, GC0015, SV1-SA, SR1-ME1077, MG0482, VF0384, and CM18. P values of the difference between CLL cells treated with miR-146b-5p vs miR CTR inhibitors or exposed to CD40L-TC vs mock cells are indicated (Wilcoxon test). ∗∗P = .0078. (C) Time course experiments to determine cell viability, cell activation, and miR-146b-5p expression following coculture with CD40L-TC. The top panels show CLL cells morphology based on their side scatter-A and forward scatter-A features before (T0) and after CD40L engagement in the representative CLL case GE1-CC190. The gate (live gate) indicates viable cells. Lower panels summarize data of the experiments in 3 different CLL cases (GE1-GA191, GE1-CC190, and SR1-ME1077). Cell viability was measured by flow cytometry by excluding annexin-V/PI-positive cells (left panel). Expression of the CD80 activation marker by purified CLL cells was evaluated by flow cytometry and is indicated as the percentage of positive cells in the viable cell gate (middle panel). The lower right panel shows miRNA expression by RT-qPCR. Data are expressed as ΔCT of miR-146b-5p vs miR-93. Data are plotted as mean ± SD. (D) RT-qPCR analysis of miR-146b 5p (CLL cases PF0024, HG0135, SR0112, CA0058, RD0468, and LG0337) and IL-12Rβ1 mRNA (CLL cases PF0024, HG0135, CA0058, RD0468, MA0151, and AR0090) expression by CLL cells transfected with miR-146b-5p inhibitors and subsequently cultured with CD40L-TC for 48 hours. miR-146b-5p expression was calculated as fold change compared with values observed in CLL cells transfected with miR-CTR inhibitors normalized to RNU44 and U6 small nuclear RNA(left panel). IL-12Rβ1 mRNA expression was calculated as fold change using CLL cells transfected with miR CTR inhibitor cells as calibrator normalized vs POL2RA gene mRNA (right panel). Data are plotted as mean ± SD. (E) IL-12Rβ1 chain expression by CLL cells from cases PF0024, HG0135, SR0112, CA0058, DF0319, RD0468, MA0151, AR0090, PD0164, and SR1-ME1077 transfected with the indicated miR inhibitors and cultured for 72 hours in the presence of CD40L-TC. Cells were analyzed by flow cytometry, and data are expressed as a percentage of positive cells. (F) IL-23R complex expression by CLL cells of the same cases analyzed in (E). Data are expressed as a percentage of positive cells (mean ± SD). P values of the difference between stimulated CLL cells and control samples are indicated (Wilcoxon test). ∗P < .05. (G) Flow cytometric analysis of IL-21 receptor expression by CLL cells treated as in (E) in a representative case DF0319. (H) Comparison of IL-23R complex or IL-21R expression by CLL cells from 5 cases (HG0135, SR0112, DF0319, RD0468, and PD0164) treated with the indicated miR inhibitors and cultured with CD40L-TC cells. Data are expressed as a percentage of positive cells (mean ± SD).
Downregulation of miR-146b-5p expression by CLL cells cocultured with CD40L-TC. (A) Comparison of IL-23R complex expression by cells from a representative CLL case (GE1-AG114) transfected with the indicated miR inhibitors or cocultured with CD40L-TC or mock control cells for 48 hours and analyzed by flow cytometry. (B) Summary of data on IL-23R complex expression (determined by flow cytometry) after a 48-hour exposure of purified CLL cells to miR-146b-5p inhibitor or CD40L-TC as in (A). CLL cells were from cases GE1-AG114, GE1-DM210, GC0015, SV1-SA, SR1-ME1077, MG0482, VF0384, and CM18. P values of the difference between CLL cells treated with miR-146b-5p vs miR CTR inhibitors or exposed to CD40L-TC vs mock cells are indicated (Wilcoxon test). ∗∗P = .0078. (C) Time course experiments to determine cell viability, cell activation, and miR-146b-5p expression following coculture with CD40L-TC. The top panels show CLL cells morphology based on their side scatter-A and forward scatter-A features before (T0) and after CD40L engagement in the representative CLL case GE1-CC190. The gate (live gate) indicates viable cells. Lower panels summarize data of the experiments in 3 different CLL cases (GE1-GA191, GE1-CC190, and SR1-ME1077). Cell viability was measured by flow cytometry by excluding annexin-V/PI-positive cells (left panel). Expression of the CD80 activation marker by purified CLL cells was evaluated by flow cytometry and is indicated as the percentage of positive cells in the viable cell gate (middle panel). The lower right panel shows miRNA expression by RT-qPCR. Data are expressed as ΔCT of miR-146b-5p vs miR-93. Data are plotted as mean ± SD. (D) RT-qPCR analysis of miR-146b 5p (CLL cases PF0024, HG0135, SR0112, CA0058, RD0468, and LG0337) and IL-12Rβ1 mRNA (CLL cases PF0024, HG0135, CA0058, RD0468, MA0151, and AR0090) expression by CLL cells transfected with miR-146b-5p inhibitors and subsequently cultured with CD40L-TC for 48 hours. miR-146b-5p expression was calculated as fold change compared with values observed in CLL cells transfected with miR-CTR inhibitors normalized to RNU44 and U6 small nuclear RNA(left panel). IL-12Rβ1 mRNA expression was calculated as fold change using CLL cells transfected with miR CTR inhibitor cells as calibrator normalized vs POL2RA gene mRNA (right panel). Data are plotted as mean ± SD. (E) IL-12Rβ1 chain expression by CLL cells from cases PF0024, HG0135, SR0112, CA0058, DF0319, RD0468, MA0151, AR0090, PD0164, and SR1-ME1077 transfected with the indicated miR inhibitors and cultured for 72 hours in the presence of CD40L-TC. Cells were analyzed by flow cytometry, and data are expressed as a percentage of positive cells. (F) IL-23R complex expression by CLL cells of the same cases analyzed in (E). Data are expressed as a percentage of positive cells (mean ± SD). P values of the difference between stimulated CLL cells and control samples are indicated (Wilcoxon test). ∗P < .05. (G) Flow cytometric analysis of IL-21 receptor expression by CLL cells treated as in (E) in a representative case DF0319. (H) Comparison of IL-23R complex or IL-21R expression by CLL cells from 5 cases (HG0135, SR0112, DF0319, RD0468, and PD0164) treated with the indicated miR inhibitors and cultured with CD40L-TC cells. Data are expressed as a percentage of positive cells (mean ± SD).
Next, purified CLL cells were transfected with the miR-146b-5p inhibitor or to the miR-CTR inhibitor for 6 hours, stimulated with CD40L-TC for 48 hours, and the concentrations of IL-12Rβ1 mRNA determined by RT-qPCR. Preexposure to the miR-146b-5p inhibitor caused a substantial increase of intracellular IL-12Rβ1 mRNA (average increase of 46.5 ± 40% [mean ± SD]) compared with the control samples (Figure 4D) irrespective of the baseline values of miR-146b-5p expression and with a wide variability depending on the propensity to CD40L activation of the different CLL clones.50 Flow cytometry tests confirmed these observations. Following pretreatment with the miR-146b-5p inhibitor, there was a consistent increase of IL-12Rβ1 (average increase of 25.6 ± 14.6% positive cells [mean ± SD]) and IL-23R complex expression (average increase of 36.4 ± 11.5% [mean ± SD]) compared with control samples (Figure 4E-F). Notably, pretreatment of the purified CLL cells with miR-146b-5p inhibitor before coculturing with CD40L-TC did not cause upregulation of the IL-21R,64 indicating a selective regulation of the miR-146b-5p on IL-23R complex expression (Figure 4G-H).
Downregulation of IL-12Rβ1 chain by enforced expression of miR-146b-5p
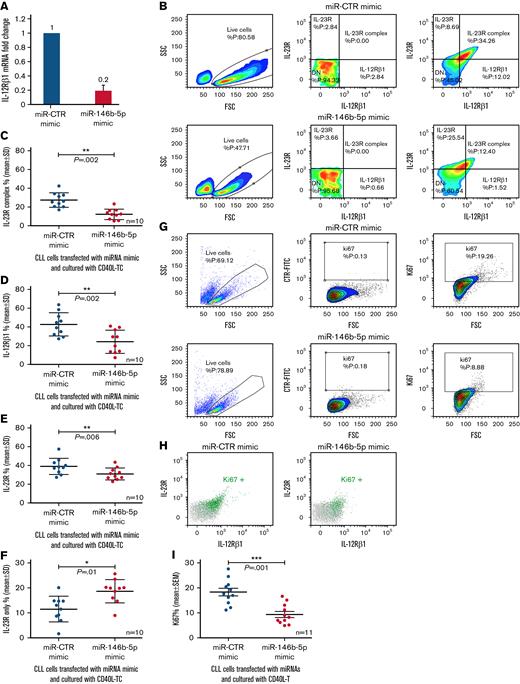
If miR-146b-5p concentrations regulate the IL-12Rβ1 expression, then a forced increase of intracellular miR-146b-5p should prevent the expression of IL-12Rβ1 following coculture with CD40L-TC. To test this, purified CLL cells from 10 different cases with different baseline miR-146b-5p expression (Figure 5 and supplemental Table 2) were cultured with miR-146b-5p mimic or miR-CTR mimic for 6 hours, CD40L-TC was added, and the cultures continued for 48 hours. Following transfection with miR-146b-5p mimics, lower IL-12Rβ1 mRNA concentrations were detected by RT-qPCR (Figure 5A). Flow cytometry confirmed that transfection with miR-146b-5p mimic prevented the expression of the surface IL-23R complex expression mediated by CD40L activation (average decrease of 55.4 ± 18% [mean ± SD]) (Figure 5B-C) mainly caused by surface downregulation of IL-12Rβ1 side chain (average decrease of 45.2 ± 22 [mean ± SD]) (Figure 5D). Concomitantly, there was a slight decrease in the overall expression of the IL-23R chain (average decrease of 19 ± 16 [mean ± SD] (Figure 5E), while the CLL cells expressing the IL-23R side chain only were increased (average increase of 39 ± 28 [mean ± SD]) (Figure 5F). Finally, a 50 ± 11.5% (mean ± SD) decrease of Ki67+ cells (Figure 5G,I) and of Ki67+ cells expressing the IL-23R complex (Figure 5H) compared with control samples was observed. Pretreatment of the purified CLL cells with miR-146b-5p mimic before coculture with CD40L-TC failed to cause upregulation of the IL-21R, as shown in supplemental Figure 9, confirming a selective effect of the miR-146b-5p mimic.
Influence of miR-146b-5p concentrations on the expression of the IL-12Rβ1 chain by CLL cells cocultured with CD40L-TC. (A) Purified CLL cells were transfected with the indicated miRNA mimics and cultured for 48 hours with CD40L-TC and IL-12Rβ1 mRNA concentrations determined by RT-qPCR. The cases studied were PF0024, DF0319, SR0112, CA0058, RD0468, MA0151, AR0090, LG0337, PD0164, and HG0135. Data are plotted as mean ± SD. (B) Evaluation of IL-23R and IL-12Rβ1 chain expression by flow cytometry in 1 representative CLL case (SR1-ME1077) cultured with CD40L-TC for 72 hours. Only viable cells were gated, and double-positive cells were considered IL-23R complex-positive. (C) IL-23R complex expression by CLL cells (cases PF0024, HG0135, SR0112, CA0058, DF0319, RD0468, MA0151, AR0090, PD0164, and SR1-ME1077) transfected with the indicated miRNAs and cultured with CD40L-TC for 72 hours. Data are expressed as a percentage of double (IL-12Rβ1+IL-23R+) positive cells (mean ± SD). (D) IL-12Rβ1 side chain, (E) IL-23R side chain, and (F) IL-23 side chain-only expression by the CLL cells from the same cases analyzed in (C). Data are expressed as a percentage of positive cells (mean ± SD). (G) Ki67 expression was analyzed by flow cytometry in CLL cells cultured with CD40L-TC for 72 hours following transfection of the indicated miRNA mimics. Data from a representative case (CLL RD0468) are shown. (H) Multiparametric flow cytometry test to detect cells expressing both IL-23R complex and Ki67 in the same representative experiment as in (E). The cells were stained for Ki67 and the IL-12Rβ1 and IL-23R chains of the IL-23 receptor complex. Ki67+ cells gated in (E) are highlighted by the green dots, whereas the whole cell population is indicated by gray dots. (I) Summary of Ki67+ cells determinations obtained on 11 different CLL cases (PF0024, HG0135, SR0112, CA0058, DF0319, RD0468, MA0151, AR0090, PD0164, LG0337, and SR1-ME1077) after a 72-hour culture with CD40L-TC following transfection of the indicated miRNA mimics. Data are expressed as mean ± SD. P values of the difference between treated and control cells are indicated (Wilcoxon test). ∗P < .05.
Influence of miR-146b-5p concentrations on the expression of the IL-12Rβ1 chain by CLL cells cocultured with CD40L-TC. (A) Purified CLL cells were transfected with the indicated miRNA mimics and cultured for 48 hours with CD40L-TC and IL-12Rβ1 mRNA concentrations determined by RT-qPCR. The cases studied were PF0024, DF0319, SR0112, CA0058, RD0468, MA0151, AR0090, LG0337, PD0164, and HG0135. Data are plotted as mean ± SD. (B) Evaluation of IL-23R and IL-12Rβ1 chain expression by flow cytometry in 1 representative CLL case (SR1-ME1077) cultured with CD40L-TC for 72 hours. Only viable cells were gated, and double-positive cells were considered IL-23R complex-positive. (C) IL-23R complex expression by CLL cells (cases PF0024, HG0135, SR0112, CA0058, DF0319, RD0468, MA0151, AR0090, PD0164, and SR1-ME1077) transfected with the indicated miRNAs and cultured with CD40L-TC for 72 hours. Data are expressed as a percentage of double (IL-12Rβ1+IL-23R+) positive cells (mean ± SD). (D) IL-12Rβ1 side chain, (E) IL-23R side chain, and (F) IL-23 side chain-only expression by the CLL cells from the same cases analyzed in (C). Data are expressed as a percentage of positive cells (mean ± SD). (G) Ki67 expression was analyzed by flow cytometry in CLL cells cultured with CD40L-TC for 72 hours following transfection of the indicated miRNA mimics. Data from a representative case (CLL RD0468) are shown. (H) Multiparametric flow cytometry test to detect cells expressing both IL-23R complex and Ki67 in the same representative experiment as in (E). The cells were stained for Ki67 and the IL-12Rβ1 and IL-23R chains of the IL-23 receptor complex. Ki67+ cells gated in (E) are highlighted by the green dots, whereas the whole cell population is indicated by gray dots. (I) Summary of Ki67+ cells determinations obtained on 11 different CLL cases (PF0024, HG0135, SR0112, CA0058, DF0319, RD0468, MA0151, AR0090, PD0164, LG0337, and SR1-ME1077) after a 72-hour culture with CD40L-TC following transfection of the indicated miRNA mimics. Data are expressed as mean ± SD. P values of the difference between treated and control cells are indicated (Wilcoxon test). ∗P < .05.
Since miR146b-5p is known to repress TRAF6 and IRAK-1, which play critical roles in NF-kB activation,27,28,31 we investigated whether enforced expression of this miRNA caused downregulation of these targets in CLL cells. Since miR146a-5p has similar effects,31 the 2 miRNAs were tested in parallel. CLL cells were exposed to miR-146a-5p or miR-146b-5p or miR-CTR mimics for 6 hours in vitro and subsequently cocultured with CD40L-TC or mock cells for 48 hours. As shown in supplemental Figure 10A-B, TRAF6 protein was downregulated in CLL cells transfected with either miR-146a-5p or miR-146b-5p mimics compared with the control samples. Some TRAF6 inhibition, although at lower concentrations, was observed in control cocultures with mock cells. Similar results were obtained when IRAK1 expression was tested by flow cytometry in the same culture settings (supplemental Figure 10C-D).
Inhibition of CLL cell growth by miR-146b-5p mimic in vivo
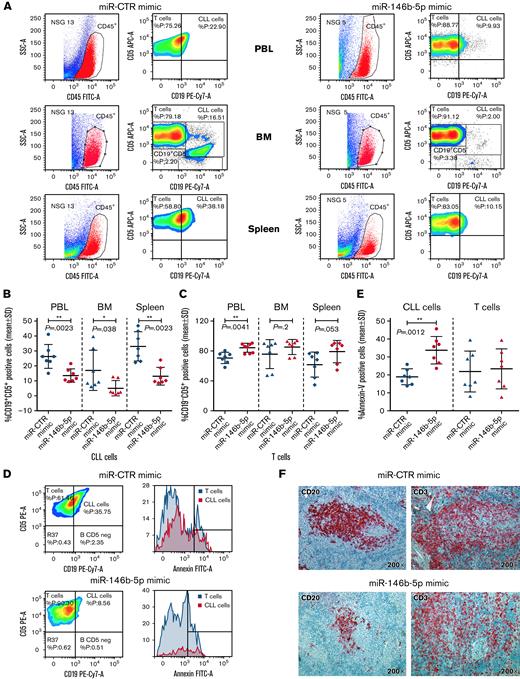
Cells from GE1-PM129 and GE1-RO148 CLL cases were cocultured with activated autologous T cells and used to generate xenografts in 10 and 4 NSG mice, respectively. After 4 to 6 weeks, all mice presented circulating human (CD45+CD19+CD5+) cells indicative of successful engraftment. The mice were subdivided into equal groups, and each group of animals was treated with either miR-146b-5p mimic or miR-CTR mimic (1 injection on alternate days for a total of 3 injections). Flow cytometry analyses of samples from peripheral blood, bone marrow, and spleen cells, 3 days after the last miRNA injection, revealed that mice treated with miR-CTR mimic had higher percentages of CD45+CD19+CD5+ CLL cells and lower percentages of CD45+CD19−CD5+ T cells than mice treated with miR-146b-5p mimic (Figure 6A-C and supplemental Table 7). Mice treated with miR-146b-5p mimic presented a higher percentage of apoptotic (annexin-V–positive) CLL cells in the spleen (Figure 6D-E). This finding was consistent with the in situ immunohistochemical (IHC) analysis showing a decrease of spleen infiltration by leukemic (human CD20+) cells after treatment with miR-146b-5p mimics (Figure 6F). Autologous T cells (human CD3+ cells), surrounding remnants of CLL infiltration foci, were still present (Figure 6E-F and supplemental Table 7). In addition, the boundaries of the follicles appeared less evident and were often disrupted by the accumulation of T cells (Figure 7A-B). Engraftment was measured by determining an IHC index derived from the combination of size and numbers of CD20+ follicles in the spleen15 (supplemental Methods). A significantly lower IHC index was observed in mice treated with the miR-146b-5p mimic compared with control mice (127.6 ± 51 vs 34 ± 31.4 [mean ± SD]; P = .007) (supplemental Table 7). Moreover, in mice treated with the miR-146b-5p mimic, there were fewer Ki67+ cells in the spleen infiltrates (Figure 7C). Double-marker IHC confirmed the presence of fewer cycling CLL cells (human Ki67/CD20+) in the spleen infiltrates of mice treated with the miR-146b-5p mimic compared with the control samples (Figure 7D).
Effects of in vivo treatment with miRNA mimics on CLL cells engrafted in NSG mice. Mice were injected with 50 × 106 CLL cells prestimulated with autologous activated T cells. After 4 to 6 weeks, blood samples were evaluated for the presence of circulating leukemic cells by flow cytometry, and after CLL cell engraftment was achieved, the mice were treated with 3 doses of miR-146b-5p or miR CTR mimic. Mice were sacrificed after 3 days from the last inoculum, and cell suspensions from the spleen, bone marrow (BM), and peripheral blood (PBL) were analyzed by flow cytometry for the percentage of CD19+CD5+ CLL cells or CD19−CD5+ T cells over the total of human CD45+ cells. (A) Flow cytometry analysis of 2 representative mice injected with CLL GE1-PM129 and treated with miR CTR mimic (NSG 13) or miR-146b-5p (NSG 5). (B) Summary of the flow cytometry analyses of 14 mice injected with CLL cells from 2 different cases (GE1-PM129 and GE1-RO148) and treated as in (A). Percentages of CD19+CD5+ CLL cells or (C) CD19−CD5+ T cells are shown. (D) Apoptotic neoplastic cells and T cells in the spleen from a miR-146b-5p (NSG 16) and a miR CTR (NSG 13) mimic-treated mouse. Apoptotic cells were detected by flow cytometry as annexin-V–positive cells by gating CD45+CD19+CD5+ CLL cells (solid red histogram profiles) or CD45+CD19−CD5+ T cells (solid blue histogram profiles). (E) Summary of the flow cytometry tests carried out in 14 mice treated as in (D). Percentages of apoptotic CLL and T cells are expressed as mean ± SD. (F) High-power magnification (200×) images of longitudinal sections of paraffin-embedded mouse spleen stained with αCD20-Ab or αCD3-Ab showing the infiltrating foci present in mice treated with miR CTR (NSG 13) or miR-146b-5p mimics (NSG 16).
Effects of in vivo treatment with miRNA mimics on CLL cells engrafted in NSG mice. Mice were injected with 50 × 106 CLL cells prestimulated with autologous activated T cells. After 4 to 6 weeks, blood samples were evaluated for the presence of circulating leukemic cells by flow cytometry, and after CLL cell engraftment was achieved, the mice were treated with 3 doses of miR-146b-5p or miR CTR mimic. Mice were sacrificed after 3 days from the last inoculum, and cell suspensions from the spleen, bone marrow (BM), and peripheral blood (PBL) were analyzed by flow cytometry for the percentage of CD19+CD5+ CLL cells or CD19−CD5+ T cells over the total of human CD45+ cells. (A) Flow cytometry analysis of 2 representative mice injected with CLL GE1-PM129 and treated with miR CTR mimic (NSG 13) or miR-146b-5p (NSG 5). (B) Summary of the flow cytometry analyses of 14 mice injected with CLL cells from 2 different cases (GE1-PM129 and GE1-RO148) and treated as in (A). Percentages of CD19+CD5+ CLL cells or (C) CD19−CD5+ T cells are shown. (D) Apoptotic neoplastic cells and T cells in the spleen from a miR-146b-5p (NSG 16) and a miR CTR (NSG 13) mimic-treated mouse. Apoptotic cells were detected by flow cytometry as annexin-V–positive cells by gating CD45+CD19+CD5+ CLL cells (solid red histogram profiles) or CD45+CD19−CD5+ T cells (solid blue histogram profiles). (E) Summary of the flow cytometry tests carried out in 14 mice treated as in (D). Percentages of apoptotic CLL and T cells are expressed as mean ± SD. (F) High-power magnification (200×) images of longitudinal sections of paraffin-embedded mouse spleen stained with αCD20-Ab or αCD3-Ab showing the infiltrating foci present in mice treated with miR CTR (NSG 13) or miR-146b-5p mimics (NSG 16).
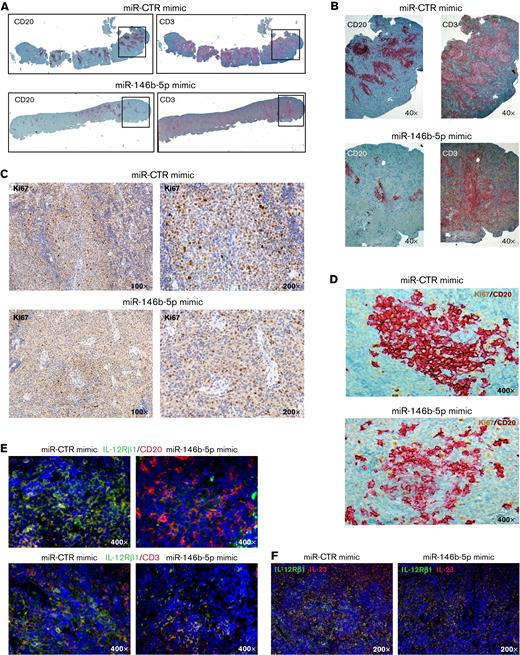
Treatment with miR-146b-5p mimic inhibits CLL growth, proliferation, and IL-12Rβ1 expression in the NSG mouse model. (A) Low magnification image (top) of longitudinal sections of paraffin-embedded mouse spleen (CLL GE1-PM129) stained for CD20 (left) or CD3 (right). The spleen from a mouse treated with miR CTR mimic (NSG13, top) or with miR-146b-5p mimic (NSG16, bottom) is represented. (B) Higher magnification (40×) of the areas of spleen highlighted by the boxes in (A) showing a decrease of neoplastic B cells (and not of T cells) in the follicular infiltrates after treatment with miR-146b-5p compared with control samples. (C) IHC analysis of Ki67+ cycling cells at 100× and 200× magnification in spleens of the same mice treated as indicated in (A). Decreased proliferating cells in the spleen infiltrate of mice treated with miR-146b-5p mimic (lower panels) compared with control samples (upper panels) is evident. (D) High magnification image (400×) of longitudinal sections of the spleens of the same mice as in (C) double-stained for CD20 and Ki67. (E) Double-marker immunofluorescence (IF) and confocal microscopy (high-magnification image 400×) analysis of IL-12Rβ1 chain and CD20 (upper panels) or CD3 chain (lower panels) and DAPI of the spleens of miR-CTR (NSG 14, left) or miR-146b-5p mimic-treated mice (NSG 5, right). (F) Double-marker IF and confocal microscopy (high-magnification 200×) analysis of IL-12Rβ1 chain and IL-23 or DAPI of a representative of the spleens of miR-CTR (NSG 14, upper panel) or miR-146b-5p mimic-treated mice (NSG 5, lower panel). IF microphotographs are representative of analyses of ≥5 low-power magnification (100×) or 10 high-power magnification (200× and 400×) microscopic fields performed on each mouse tissue sample.
Treatment with miR-146b-5p mimic inhibits CLL growth, proliferation, and IL-12Rβ1 expression in the NSG mouse model. (A) Low magnification image (top) of longitudinal sections of paraffin-embedded mouse spleen (CLL GE1-PM129) stained for CD20 (left) or CD3 (right). The spleen from a mouse treated with miR CTR mimic (NSG13, top) or with miR-146b-5p mimic (NSG16, bottom) is represented. (B) Higher magnification (40×) of the areas of spleen highlighted by the boxes in (A) showing a decrease of neoplastic B cells (and not of T cells) in the follicular infiltrates after treatment with miR-146b-5p compared with control samples. (C) IHC analysis of Ki67+ cycling cells at 100× and 200× magnification in spleens of the same mice treated as indicated in (A). Decreased proliferating cells in the spleen infiltrate of mice treated with miR-146b-5p mimic (lower panels) compared with control samples (upper panels) is evident. (D) High magnification image (400×) of longitudinal sections of the spleens of the same mice as in (C) double-stained for CD20 and Ki67. (E) Double-marker immunofluorescence (IF) and confocal microscopy (high-magnification image 400×) analysis of IL-12Rβ1 chain and CD20 (upper panels) or CD3 chain (lower panels) and DAPI of the spleens of miR-CTR (NSG 14, left) or miR-146b-5p mimic-treated mice (NSG 5, right). (F) Double-marker IF and confocal microscopy (high-magnification 200×) analysis of IL-12Rβ1 chain and IL-23 or DAPI of a representative of the spleens of miR-CTR (NSG 14, upper panel) or miR-146b-5p mimic-treated mice (NSG 5, lower panel). IF microphotographs are representative of analyses of ≥5 low-power magnification (100×) or 10 high-power magnification (200× and 400×) microscopic fields performed on each mouse tissue sample.
Furthermore, staining with specific antibodies showed fewer IL-12Rβ1–expressing cells in mice treated with miR-146b-5p mimic than in the control samples (Figure 7E-F). Notably, a consistent number of cells present in the CLL cell aggregates were stained by anti–IL-23 mAb, indicating that the miR146b-5p mimic treatment did not affect IL-23 cytokine production (Figure 7F). Likewise, there were numerous T cells in the tissues analyzed, indicating that the T-cell compartment was not prominently affected by miR-146b-5p mimic treatment (Figure 7E).
Discussion
The idea for this study stemmed from the consideration that miR-146b-5p had a relevant prognostic impact in CLL and that the IL-23/IL-23R complex loop is important for promoting CLL cell clonal expansion. Since IL-12Rβ1 is expressed following cell activation, this step may represent a relevant checkpoint for the functioning of the loop,50 and miR-146b-5p could conceivably determine the cell’s susceptibility to IL-23 by regulating IL-12Rβ1 receptor expression.
The collected evidence supports the working hypothesis: miR-146b-5p proved capable of binding to the IL-12Rβ1 chain mRNA in an in vitro luciferase assay, whereas miR-146a-5p failed despite sharing the same seed sequence. A partial explanation for this failure could be that the binding of miRNAs associated with the argonaute protein to the relevant mRNA is influenced by sequences flanking the binding sites and by additional noncanonical binding sites. Thus, small sequence variations outside the seed sequence, and the different posttranslational processing of the 2 miRNAs, may cause variations in their binding to target mRNA.65 The capacity of miR-146b-5p to regulate IL-12Rβ1 expression was confirmed by experiments with specific miR-146b-5p mimics and inhibitors because the former prevented and the latter promoted IL-12Rβ1 expression. This effect was selective given that the expression of IL-21R, which also plays an important role in regulating CLL cell expansion,64,66 was unaffected by miR-146b-5p. Notably, miR-146b-5p did not bind to IL-23 mRNA and did not appear to influence IL-23 production by CLL cells, indicating that IL-12Rβ1 chain expression is a major regulatory step in the IL-23/IL-23R complex loop.
NSG mice engrafted with CLL cells and treated with miR-146b-5p mimic presented a reduction of both circulating and tissue-infiltrating leukemic cells compared with mice treated with CTR mimics and a disruption of the leukemic nodules, which had less defined boundaries, inferior numbers of proliferating cells, and appeared infiltrated by T cells. The expression of the IL-12Rβ1 chain by leukemic cells was markedly diminished. The concentrations of human T cells remained apparently unaltered in the engrafted NSG mice upon miR-146b-5p mimic administration, indicating that the treatment did not influence T-cell viability in this setting, although previous reports described a regulatory function of miR-146b-5p in follicular Th cells and regulatory T cells.45,67 Whether the T-cell subset distribution is altered remains to be ascertained.
The activation of the IL-23/IL-23R complex loop in CLL cells is achieved mainly by stimulation through the surface CD40-dependent, not the BCR-dependent pathway.50 CD40 is a member of the TNFR family and requires interaction with TRAF6 and IRAK1 to activate NF-kB.28,29 Elevated concentrations of miR-146a-5p and of miR-146b-5p cause downregulation of IRAK1 and TRAF6 (supplemental Figure 10) in CLL, in principle, rendering NF-kB activation and stimulation via surface CD40 less effective.27,31,32 However, the observation that miR-146a-5p, which downregulates TRAF6 and IRAK1 expression as efficiently as miR-146b-5p, was not associated with prognosis in CLL suggested that the IL-23/IL-23R complex loop had a more critical role in regulating CLL cell growth. Interestingly, mice in which miR-146a-5p or miR-146b-5p is knocked out (KO) both develop lymphomas, although only the lymphomas originated in miR-146b-5p KO mice present a resemblance to human CLL.44 The reasons for this hierarchy in the mechanisms regulating CLL clonal expansion are far from clear. One possibility is that additional signals delivered by surface molecules different from CD40 and not requiring TRAF6 and IRAK1 adaptors are involved in activating the IL-23/IL-23R loop in vivo. An alternative and not mutually exclusive option could be offered by the redundancy of the TRAF/IRAK family members, whereby other molecules of the same families could substitute for the downregulation of TRAF6 and IRAK1 induced by the miR-146a/b.68,69 Notably, other miRNAs can regulate IL-23 stimulatory signals. This is the case of miR-221 and miR-222 that negatively regulate the susceptibility of Th17 cells to IL-23 stimulation by modulation of the IL-23R complex.70
The issue as to why CLL clones are heterogeneous in the miR-146b-5p concentrations is presently unclear, although it could be related to the different states of activation of the cells from the different CLL clones. This hypothesis is supported by the observations that CLL cell activation with CD40L-TC causes downregulation of miR-146b-5p concentrations in vitro and that the lowest miR-146b-5p concentrations are detected in IGHV-UM cases whose leukemic cells are at the highest activation status determined by surface marker analysis.71,72 An alternative and not mutually exclusive hypothesis poses that lesions of the miR-146b gene or regulatory DNA sequences facilitate the maintenance of low miR-146b-5p concentrations in the most aggressive CLL clones. However, this hypothesis is made unlikely by the finding that CNAs were very low in the database analysis we have carried out, and virtually no mutations of the miR-146b-5p locus are detectable in the same database.57,58 Alterations in the methylation of the miR-146b-5p locus of CLL cells compared with normal cells have been noticed and are reported in the Blueprint data analysis portal61 (supplemental Figure 5), a finding that could at least in part explain the heterogeneity of miR-146b expression in CLL, possibly dependent on the activation status of the neoplastic clones. This issue is currently being investigated.
The present study has translational relevance as it indicates miR-146b-5p is a potential target through which the susceptibility of CLL cells to IL-23 could be modified. Future studies should investigate a strategy based on increasing intracellular miR-146b-5p concentrations as an application for CLL therapy, either alone or combined with anti–IL-23 mAbs,73-75 in the attempt to eradicate CLL, which so far has proven virtually incurable.
Acknowledgments
In addition to the listed authors, the following Investigators participated in this study as part of the Gruppo Italiano Studio Linfomi (GISL): Gianni Quintana, Divisione di Ematologia, Presidio Ospedaliero “A.Perrino”, Brindisi; Giovanni Bertoldero, Dipartimento di Oncologia, Ospedale Civile, Noale, Venezia; Paolo Di Tonno, Dipartimento di Ematologia, Ospedale di Venere, Bari; Robin Foà and Francesca R Mauro, Divisione di Ematologia, Università La Sapienza, Roma; Nicola Di Renzo, Unità di Ematologia, Ospedale Vito Fazzi, Lecce; Maria Cristina Cox, Ematologia, Azienda Ospedaliera Sant’Andrea, Università La Sapienza, Roma; Stefano Molica, Dipartimento di Oncologia ed Ematologia, Pugliese-Ciaccio Hospital, Catanzaro; Attilio Guarini, Unità di Ematologia e Trapianto di Cellule Staminali, Istituto di Oncologia “Giovanni Paolo II”, Bari; Antonio Abbadessa, Unità Operativa Complessa di Oncoematologia Ospedale “S. Anna e S. Sebastiano”, Caserta; Francesco Iuliano, Unità Operativa Complessa di Oncologia, Ospedale Giannettasio, Rossano Calabro, Cosenza; Omar Racchi, Ospedale Villa Scassi Sampierdarena, Genova; Mauro Spriano, Ematologia, IRCCS Policlinico San Martino, Genova; Felicetto Ferrara, Divisione di Ematologia, Ospedale Cardarelli, Napoli; Monica Crugnola, Ematologia, CTMO, Azienda Ospedaliera Universitaria di Parma; Alessandro Andriani, Dipartimento di Ematologia, Ospedale Nuovo Regina Margherita, Roma; Nicola Cascavilla, Unità di Ematologia e Trapianto di Cellule Staminali, IRCCS Ospedale Casa Sollievo della Sofferenza, San Giovanni Rotondo; Lucia Ciuffreda, Unità di Ematologia, Ospedale San Nicola Pellegrino, Trani; Graziella Pinotti, Unità Operativa Oncologia Medica, Ospedale di Circolo Fondazione Macchi, Varese; Anna Pascarella, Unità Operativa di Ematologia, Ospedale dell'Angelo; Venezia-Mestre, Maria Grazia Lipari, Divisione di Ematologia, Ospedale Policlinico, Palermo; Francesco Merli, Unità Operativa di Ematologia, Azienda Ospedaliera Maria Nuova; Reggio Emilia, Luca Baldini Istituto di Ricovero e Cura a Carattere Scientifico Cà Granda-Maggiore Policlinico, Milano; Caterina Musolino, Divisione di Ematologia, Università di Messina; Agostino Cortelezzi, Ematologia and Centro Trapianti Midollo Osseo, Foundation IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milano; Francesco Angrilli, Dipartimento di Ematologia, Ospedale Santo Spirito, Pescara; Ugo Consoli, Unità Operativa Semplice di Emato-Oncologia, Ospedale Garibaldi-Nesima, Catania; Gianluca Festini, Centro di Riferimento Ematologico-Seconda Medicina, Azienda Ospedaliero-Universitaria; Ospedali Riuniti, Trieste, Giuseppe Longo, Unità di Ematologia, Ospedale San Vincenzo, Taormina; Daniele Vallisa and Annalisa Arcari, Unità di Ematologia, Dipartimento di Onco-Ematologia, Guglielmo da Saliceto Hospital, Piacenza; Francesco Di Raimondo and Annalisa Chiarenza, Divisione di Ematologia, Università di Catania Ospedale Ferrarotto, Catania; Iolanda Vincelli, Unità di Ematologia, Azienda Ospedaliera of Reggio Calabria; Donato Mannina, Divisione di Ematologia, Ospedale Papardo, Messina, Italy. For Each Institution, the Ethics Review Committee that approved the protocol is listed below. Unità di Ematologia e Trapianto di Cellule Staminali, Istituto di Oncologia “Giovanni Paolo II”, Bari comitato etico Istituto di Oncologia “Giovanni Paolo II”, di Bari, Italy; Dipartimento di Ematologia, Ospedale di Venere, Bari comitato etico Ospedale di Venere, Bari, Italy; Unità Operativa Complessa Ematologia e Trapianto, Ospedale “Mons. R. Dimiccoli” - Barletta comitato etico Ospedale “Mons. R. Dimiccoli” - Barletta, Italy Divisione di Ematologia, Presidio Ospedaliero “A. Perrino”, Brindisi comitato etico Presidio Ospedaliero “A. Perrino”, Brindisi, Italy; Unità Operativa Complessa di Oncoematologia Ospedale “S. Anna e S. Sebastiano”, Caserta comitato etico Ospedale “S. Anna e S. Sebastiano”, Caserta, Italy; Divisione di Ematologia, Università di Catania Ospedale Ferrarotto, Catania comitato etico Ospedale Ferrarotto, Catania, Italy; Unità Operativa Complessa di Emato-Oncologia, Ospedale Garibaldi-Nesima, Catania comitato etico Ospedale Garibaldi - Nesima, Catania, Italy; Dipartimento di Oncologia ed Ematologia, Pugliese-Ciaccio Hospital, Catanzaro comitato etico Pugliese-Ciaccio Hospital, Catanzaro, Italy; Unità Operativa Complessa di Ematologia, Azienda Ospedaliera Cosenza comitato etico Azienda Ospedaliera Cosenza, Italy; Unità Operativa Complessa di Oncologia, Ospedale Giannettasio, Rossano Calabro, Cosenza comitato etico Ospedale Giannettasio, Rossano Calabro, Italy; Divisione di Ematologia, Ospedale Policlinico, Palermo, comitato etico Ospedale Policlinico, Palermo, Italy; Ematologia ospedale Goretti, Latina comitato etico ospedale Goretti, Latina, Italy, Clinica Ematologica, DIMI, Genova, comitato etico IRCCS Ospedale Policlinico San Martino, Genova, Italy; Oncologia medica C IRCCS Ospedale Policlinico San Martino, Genoa, Italy comitato etico IRCCS Ospedale Policlinico San Martino, Genova, Italy; Ospedale Villa Scassi Sampierdarena, Genova comitato etico Ospedale Villa Scassi, Genova, Italy; Ematologia, Azienda Ospedaliera San Martino, Genova comitato etico IRCCS Policlinico San Martino, Genova, Italy; Unità di Ematologia, Ospedale Vito Fazzi, Lecce comitato etico Ospedale Vito Fazzi, Lecce, Italy; Unità Operativa Complessa di Ematologia Ospedale di Matera comitato etico Ospedale di Matera, Italy; Divisione di Ematologia, Ospedale Papardo, Messina, Italy comitato etico Ospedale Papardo, Messina, Italy; Divisione di Ematologia, Università di Messina comitato etico Ospedale di Messina, Italy; Ematologia and Centro Trapianti Midollo Osseo, Foundation IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milano comitato etico Ospedale Maggiore Policlinico di Milano, Italy; Dipartimento di Oncologia, Ospedale Civile, Noale, Venezia comitato etico Ospedale Civile, Noale, Italy; Oncoematologia Policlinico di Modena comitato etico provinciale di Modena Italy; Divisione di Ematologia, Ospedale Cardarelli, Napoli comitato etico Ospedale Cardarelli, Napoli, Italy; Ematologia, Centro Trapianti Midollo Osseo, Azienda Ospedaliera Universitaria di Parma comitato etico Azienda Ospedaliera Universitaria di Parma, Italy; Dipartimento di Ematologia, Ospedale Santo Spirito, Pescara comitato etico Ospedale Santo Spirito, Pescara, Italy; Unità di Ematologia, Dipartimento di Onco-Ematologia, Guglielmo da Saliceto Hospital, Piacenza comitato etico Guglielmo da Saliceto Hospital, Piacenza, Italy; Unità di Ematologia, Azienda Ospedaliera of Reggio Calabria comitato etico Azienda Ospedaliera of Reggio Calabria, Italy; Unità Operativa di Ematologia, Azienda Ospedaliera Maria Nuova, Reggio Emilia comitato etico Azienda Ospedaliera Maria Nuova, Reggio Emilia, Italy; Unità Operativa Complessa di Ematologia e Trapianto di Cellule Staminali IRCCS-CROB di Rionero in Vulture, Potenza comitato etico IRCCS-CROB di Rionero in Vulture, Italy; Dipartimento di Ematologia, Ospedale Nuovo Regina Margherita, Roma comitato etico Ospedale Nuovo Regina Margherita, Roma, Italy; Ematologia, Azienda Ospedaliera Sant’Andrea, Università La Sapienza, Roma comitato etico Azienda Ospedaliera Sant’Andrea, Roma, Italy; Divisione di Ematologia, Università La Sapienza, Roma comitato etico Università La Sapienza, Roma, Italy; Unità di Ematologia e Trapianto di Cellule Staminali, IRCCS Ospedale Casa Sollievo della Sofferenza, San Giovanni Rotondo comitato etico IRCCS Ospedale Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy; Unità di Ematologia, Ospedale San Vincenzo, Taormina, comitato etico Ospedale San Vincenzo, Taormina, Italy; Unità di Ematologia, Ospedale San Nicola Pellegrino, Trani comitato etico Ospedale San Nicola Pellegrino, Trani, Italy; Centro di Riferimento Ematologico-Seconda Medicina, Azienda Ospedaliero-Universitaria, Ospedali Riuniti, Trieste comitato etico Ospedali Riuniti, Trieste, Italy; Unità Operativa Oncologia Medica, Ospedale di Circolo Fondazione Macchi, Varese comitato etico Ospedale di Circolo Fondazione Macchi, Varese, Italy; Unità Operativa di Ematologia, Ospedale dell'Angelo, Venezia-Mestre comitato etico Ospedale dell'Angelo, Venezia-Mestre, Italy; IRCCS Cà Granda-Maggiore Policlinico, Milano comitato etico Policlinico, Milano, Italy.
The primers and NOTCH1 and TP53 mutations assessment were kindly provided by TIB Molbiol srl (Genoa, Italy). The authors thank Fabio Ghiotto for helpful support in the graphical elaboration of the visual abstract.
This work was supported by Associazione Italiana Ricerca sul Cancro (AIRC) Grant 5 × mille ID.9980, (to M.F., F.M., and A.N.); AIRC I.G. ID.14326 (to M.F.), ID.15426 (to F.F.), ID.22145 (to C.T.), ID.16722 (to A.N.), ID.24365 (to A.N.); AIRC Accelerator Award ID. 24296 (to C.T.); AIRC and Fondazione CaRiCal cofinanced Multi-Unit Regional Grant 2014 n.16695 (to F.M.); Italian Ministry of Health 5 × 1000 funds 2014 (to G.C. and. A.I.), 2015 (to F.F. and G.F.) and 2016 (to F.F., G.F., and G.C.); Italian Ministry of Health (Ricerca Corrente); Compagnia S. Paolo, Turin, Italy, Project 2017.0526 (to G.F.). Associazione Italiana contro le Leucemie-Linfomi e Mieloma (AIL, Cosenza, to F.F.); Gilead fellowship program 2016 (M.C.) and 2017 (G.C.).
Authorship
Contribution: S.M., A.G.R., F.F., F.M., M. Ferrarini., and G. Cutrona. designed the study; S.M., A.G.R., M. Colombo, M. Cardillo, M. Fabbi, K.T., R.M., D.R., G. Cerruti, S.B., S.S., P.B., S.P., S.F., P. Monti, P. Menichini, G.F., and V.C. performed the experiments in vitro; S.M., L.E., and M. Cilli conducted and supervised the experiments in vivo; F.T., A.C., and K.T. performed the analyses on metadata for mutations and CNA and epigenome modifications of the miR146b-5p locus; F.M. and G. Cutrona performed the statistical analysis; C.T. and R.F. performed the histopathological analysis; F.M., E.A., A.I., M.F., and M.G. provided patient samples and clinical expertise; M. Ferrararini and G. Cutrona wrote the manuscript, F.M., A.N., C.T., F.F., and A.G.R. reviewed the manuscript; and all authors analyzed data and approved the final version of the manuscript.
Conflict-of-interest disclosures: The authors declare no competing financial interests.
Correspondence: Giovanna Cutrona, Molecular Pathology Unit, IRCCS-Ospedale Policlinico San Martino, L.go Rosanna Benzi, 10-16132 Genoa, Italy; e-mail: giovanna.cutrona@hsanmartino.it; and Franco Fais, Department of Experimental Medicine, University of Genoa, Via De Toni, 14 16132 Genoa, Italy; e-mail: franco.fais@unige.it.
References
Author notes
The data are deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE40533. Contact the corresponding author for other forms of data sharing: giovanna.cutrona@hsanmartino.it.
The full-text version of this article contains a data supplement.
final version published online 13 October 2022
S.M. and A.G.R. are joint first authors.
F.M., M. Ferrarini, and G.C. are joint last authors.