Key Points
High rates of bloodstream infection (BSI) were seen after fecal microbiota transplantation (FMT) in patients with graft-versus-host disease.
Metagenomic analysis pointed to recipients’ indigenous microbiota as the BSI source, ruling out direct involvement of FMT donations.
Abstract
We observed high rates of bloodstream infections (BSIs) following fecal microbiota transplantation (FMT) for graft-versus-host-disease (33 events in 22 patients). To trace the BSIs' origin, we applied a metagenomic bioinformatic pipeline screening donor and recipient stool samples for bacteremia-causing strains in 13 cases. Offending strains were not detected in FMT donations. Enterococcus faecium, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii could be detected in stool samples before emerging in the blood. In this largest report of BSIs post-FMT, we present an approach that may be applicable for evaluating BSI origin following microbiota-based interventions. Our findings support FMT safety in immunocompromised patients but do not rule out FMT as an inducer of bacterial translocation.
Introduction
Acute graft-versus-host disease (GVHD) is a major cause of allogeneic hematopoietic stem cell transplantation (HSCT)–related mortality.1 Donor alloreactive lymphocytes attack host tissue recognized as foreign by the new immune system.2 Importantly, the gastrointestinal (GI) microbiome modulates GVHD.3,4 Immunosuppression with glucocorticosteroids is the frontline treatment of acute GVHD. Nonresponders are treated with additional immunosuppressants, which are only partially effective and increase infection risk.2 Preliminary studies suggest that modifying the host microbiota through fecal microbiota transplantation (FMT) is an effective and safe therapy for steroid-resistant intestinal acute GVHD.5-9 However, DeFilipp et al10 reported 2 bloodstream infection (BSI) cases of extended-spectrum β-lactamase–producing Escherichia coli after FMT in immunocompromised patients; the infecting strain was traced to the FMT donor’s stool.
We noticed high rates of BSI in a pilot study involving FMT treatment of steroid-resistant and steroid-dependent GVHD. This led us to investigate the BSI source and whether FMT introduced offending bacterial strains.
Methods
Fecal microbiota transplants were administered as frozen capsules as part of an extension of a nonrandomized pilot study (registered at www.clinicaltrials.gov as #NCT03214289) evaluating the safety and efficacy of FMT in steroid-resistant or steroid-dependent acute GVHD. Steroid-resistant lower GI acute GVHD was defined as intestinal GVHD that did not improve within 7 days after initial steroid therapy (≥1 mg/kg of methylprednisolone) or progressed after 5 days of treatment. Steroid dependency was defined as recurrence of lower GI acute GVHD during steroid tapering.11
Blood cultures were performed in patients experiencing spiking fever or presenting with signs of hemodynamic instability. The exception was in patients receiving IV total parenteral nutrition, where daily surveillance cultures were performed per institutional protocol.
Microbial analysis of stool and fecal microbiota transplant samples and blood culture isolates was performed using 16S ribosomal RNA and shotgun metagenomic sequencing. The 16S ribosomal RNA gene sequence data reported in this article have been deposited in the European Bioinformatics Institute database (accession number ERP132638). The shotgun sequence raw data was deposited in the SRA-NCBI database under the BioProject number PRJNA764525. The study was approved by the Chaim Sheba Medical Center Institutional Review Board and was performed according to the Declaration of Helsinki.
Results and discussion
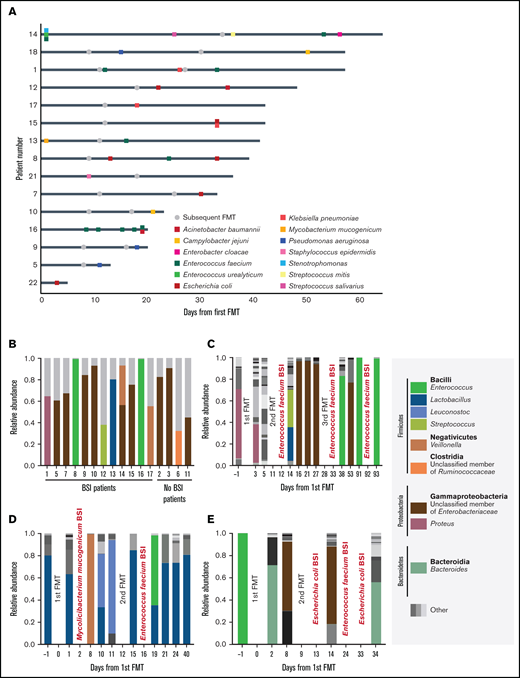
Between 2017 and 2019, we treated 22 patients with steroid-refractory and steroid-dependent (n = 21 and 1, respectively) GI acute GVHD undergoing FMT (supplemental Table 1). Before FMT, participants were receiving a median methylprednisolone dose of 1.6 mg/kg (interquartile range, 1-2); additional GVHD treatments included mycophenolate mofetil (n = 22 of 22), budesonide (n = 22 of 22), cyclosporine (n = 21 of 22), and ruxolitinib (n = 3 of 22). Patients received a course of 30 frozen fecal microbiota transplant capsules over 2 consecutive days. FMT could be repeated at the treating physician’s discretion. Overall, 44 FMTs were performed (7, 8, and 7 participants underwent 1, 2, and 3 courses of FMT, respectively). In total, 33 BSIs, occurring up to 30 days after the last FMT, were observed in 15 (68%) of 22 patients. The median time from FMT to BSI was 7 days (interquartile range, 4-19; supplemental Table 2). Enterococcus faecium (10 events) and E coli (6 events) were the most prevalent BSI-causing strains (Figure 1A; supplemental Table 3). Overall, pre-FMT patient features were similar between patients with and without BSI (supplemental Table 1), except for a longer duration between GVHD onset to FMT in patients developing a BSI (22 vs 11 days; P = .0149). Immunosuppression and increased gut permeability resulting from intestinal barrier damage,12 goblet and paneth cell dysfunction, and gut dysbiosis13,14 contributed to the high incidence of BSI in patients with acute GI GVHD. The previously reported BSI rate in patients with high-grade and steroid-resistant acute GVHD was ∼50%.15,16 The unusually high rate of BSI in our population and the proximity between the infection and FMT suggested a potentially causal association. To trace the BSI source, we characterized host and donor stool samples as well as blood culture isolates by metagenomic sequencing.
BSIs and gut bacterial composition in fecal microbiota transplant recipients. (A) Swimmer plot describing BSI events relative to FMT. Each row represents 1 patient who experienced at least 1 BSI event after FMT treatment. Day 0 is the day of the first FMT; gray dots represent subsequent FMT courses. Squares represent positive blood cultures that are color coded by species. (B) Compositional plot of dominating bacterial taxa (by 16S ribosomal RNA gene analysis) before and after FMT and BSI. Monodominating taxa are coded by color. Taxa with <30% relative abundance are summarized as other and colored gray. (C-E) Temporal dynamics of the gut microbiota (at the genus level) during FMT sessions. BSI type and timing appear in red. Patients 1 (C), 13 (D), and 8 (E). Monodominating taxa are coded by color. Taxa with a relative abundance of <30% are summarized as other.
BSIs and gut bacterial composition in fecal microbiota transplant recipients. (A) Swimmer plot describing BSI events relative to FMT. Each row represents 1 patient who experienced at least 1 BSI event after FMT treatment. Day 0 is the day of the first FMT; gray dots represent subsequent FMT courses. Squares represent positive blood cultures that are color coded by species. (B) Compositional plot of dominating bacterial taxa (by 16S ribosomal RNA gene analysis) before and after FMT and BSI. Monodominating taxa are coded by color. Taxa with <30% relative abundance are summarized as other and colored gray. (C-E) Temporal dynamics of the gut microbiota (at the genus level) during FMT sessions. BSI type and timing appear in red. Patients 1 (C), 13 (D), and 8 (E). Monodominating taxa are coded by color. Taxa with a relative abundance of <30% are summarized as other.
Intestinal bacterial monodomination (ie, bacterial relative abundance ≥30%)14 was ubiquitous across patients before the first FMT (Figure 1B), irrespective of subsequent BSI development. Indeed, dysbiosis is characteristic of acute GVHD.17 All samples collected 0 to 5 days before the BSI event were monodominated (Figure 1C-E; supplemental Figure 1). Notably, in 5 of 6 BSIs caused by E coli and 9 of 10 BSIs caused by an enterococcal strain, patient stool was dominated by the corresponding genus/family, suggesting a GI source of the BSI. In those undergoing allogeneic HSCT, intestinal monodomination by both bacteria has been shown to precede their emergence in the blood.14
Next, we sought to determine whether the strains detected in the blood culture samples originated from the fecal microbiota transplant capsules or the host’s indigenous intestinal population. We performed shotgun metagenomic sequencing in a subset of 13 of the 33 positive blood cultures, coupled with corresponding patient stool samples and donor-derived fecal microbiota transplant capsules produced from the same fecal donation. Blood culture samples sent for sequencing were selected at random. We applied a detection pipeline consisting of 3 steps (Figure 2A).18 First, the genomes of strains from blood culture samples were assembled. Next, genes were predicted and classified into species core genes and strain gene groups. Finally, metagenomic reads from fecal microbiota transplant capsules and recipient stool samples were aligned against their respective blood sample genomes. The contents of species and strain genes in each sample were evaluated and used to determine whether a sample contained a specific strain.
Tracing the source of BSI. (A) Scheme describing the BSI source detection pipeline: collection of relevant biological samples for shotgun sequencing (patient’s positive blood cultures, stool samples, and donor-derived fecal microbiota transplant capsules) (i); construction of the reference genomes using sequences of the positive blood culture strains (genome assembly and gene classification into species core genes and strain gene groups) (ii); and identification of BSI strains in the metagenomic samples (searching for species and strain genes identified in the reference genome) (iii). (B) Detection of blood culture strains in fecal samples. Rows are grouped by patients and correspond with bacterial strains detected in the blood cultures. Using the metagenomic bioinformatic pipeline, we searched for BSI-causing strains within donor and recipient stool samples. Offending bacterial strains were absent from fecal microbiota transplant donor capsules. However, they could be detected in a portion of recipients’ stool samples at various time points before and after the FMT and BSI event. Source data for the figure are available in the data supplement. aGVHD, acute GVHD. Reproduced with permission, copyright © Elvire Thouvenot.
Tracing the source of BSI. (A) Scheme describing the BSI source detection pipeline: collection of relevant biological samples for shotgun sequencing (patient’s positive blood cultures, stool samples, and donor-derived fecal microbiota transplant capsules) (i); construction of the reference genomes using sequences of the positive blood culture strains (genome assembly and gene classification into species core genes and strain gene groups) (ii); and identification of BSI strains in the metagenomic samples (searching for species and strain genes identified in the reference genome) (iii). (B) Detection of blood culture strains in fecal samples. Rows are grouped by patients and correspond with bacterial strains detected in the blood cultures. Using the metagenomic bioinformatic pipeline, we searched for BSI-causing strains within donor and recipient stool samples. Offending bacterial strains were absent from fecal microbiota transplant donor capsules. However, they could be detected in a portion of recipients’ stool samples at various time points before and after the FMT and BSI event. Source data for the figure are available in the data supplement. aGVHD, acute GVHD. Reproduced with permission, copyright © Elvire Thouvenot.
Overall, we assembled 14 genomes from the 13 blood culture samples. E coli was detected in 4 samples, 2 of which were from the same patient and represented the same strain. Additional bacterial strains identified were E faecium (4 genomes, 2 of which represented the same strain in the same patient), Pseudomonas aeruginosa, Brevibacterium frigoritolerans, Acinetobacter baumannii, Mycobacterium mucogenicum, and Campylobacter jejuni (1 genome each). All the genomes were estimated to be >95% complete and <2% contaminated (supplemental Table 2). Assembled genomes were concordant with positive blood culture isolates. However, in patient 8, E faecium and B frigoritolerans were both identified by genome assembly, whereas only E faecium grew in the blood culture. B frigoritolerans has been previously isolated from soil and is therefore likely a contaminant.19
We mapped the reads from the fecal microbiota transplant capsules and patient stool samples against the assembled bacterial genomes from the blood cultures (supplemental Table 4). No BSI-related bacterial strains were detected in the fecal microbiota transplant capsules (supplemental Table 4). Notably, the analysis resulted in inconclusive results for 3 fecal microbiota transplant capsules because of marginal coverage of the target genomes. Resequencing at ∼3.5 to 5.5 times the original depth confirmed that the BSI strains were not present at >0.01% community relative abundance threshold. The blood culture strains were identified in 7 of 13 BSI cases (Figure 2B). There were 4 stool samples in which results were inconclusive (data supplement). The BSI-causing bacterial strains could be detected at a low relative abundance up to 38 days before the BSI, as shown in the patient with A baumannii infection. Overall, our findings support the host intestinal microbiota as a reservoir for both environmentally acquired microbes and normally commensal symbionts causing infection after translocation.20
Evidence supporting a mechanistic link between microbiota and GVHD has led investigators to modulate the host gut microbial composition using FMT.3,4 Several groups have reported a favorable safety profile with FMT, including in immunocompromised states such as GVHD.5-9 High-grade acute GI GVHD is an established risk factor for BSI.16 Because our trial was a single-arm study, we could not determine if the high BSI rates were caused, directly or indirectly, by the intervention.
On the basis of the metagenomic-based strain detection method results, we conclude that it is unlikely that the fecal microbiota transplant capsules were the direct source of BSI. The strain-detection procedure we used enables the accurate identification of a target genome given 0.4× coverage. This coverage is lower than that required by single-nucleotide variation–based methods such as StrainPhlAn, which requires at least 2× coverage.21 For samples in the current study, the 0.4× coverage limit roughly corresponds to 0.1% to 0.2% relative abundance in the community. False positives resulting from the presence of multiple strains of the target genome species are unlikely, because at least 7 strains at sufficient coverage are required (data supplement).
Profound immunosuppression secondary to active GVHD, drugs, poor nutrition, and prolonged hospitalization may contribute to BSI development. Furthermore, these factors may entail a selective advantage to pathobionts colonizing the gut well before translocating to blood, as detected in our analysis.14 FMT has been shown to promote immune reconstitution in those undergoing allogeneic HSCT.22 Nevertheless, the indirect involvement of FMT in propagating bacterial translocation from the gut through the damaged intestinal barrier, possibly through a local gut mucosal inflammatory reaction, cannot be ruled out.23 To our knowledge, our study represents the most extensive effort to track the source of BSI after FMT. Given the current literature, microbiome therapeutics, such as FMT, remain a viable option for treatment and prevention of GVHD and should be studied further in larger cohorts with more frequent blood and stool sampling. However, we argue that future trials testing microbiota-based approaches should use a similar genomic strategy to determine whether infectious adverse events are directly related to the intervention.
Acknowledgments
The authors thank Elvire Thouvenot-Nitzan for figure design and illustration. R.S. is supported by the American Society of Hematology Fellow Scholar Award.
This work was supported by the Dahlia Greidinger Anti Cancer Fund, the Gasner Fund for Medical Research, and an institutional grant from the Chaim Sheba Medical Center.
Authorship
Contribution: A.E, I.S., I.Y., O.K., and R.S. planned the study; A.E, I.S., J.A.F, D.B., and R.S. analyzed the data; R.S, I.D., A.N., M.G., I.H., A.S., and T.Z. enrolled patients; I.Y. manufactured the fecal microbiota transplant capsules; M.G. coded data into the database; A.E., I.S., and R.S. wrote the manuscript; A.E., A.N., O.K., and R.S. supervised the analysis and study; and all authors edited and reviewed the manuscript.
Conflict-of-interest disclosure: I.Y. is a medical advisor to Mybiotics Ltd. The remaining authors declare no competing financial interests.
Correspondence: Roni Shouval, Adult Bone Marrow Transplant Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, 1275 York Ave, Box 298, New York, NY 10065; e-mail: shouval@gmail.com; and Omry Koren, Azrieli Faculty of Medicine, Bar Ilan University, 8 Henrietta Szold St, Safed, Israel; e-mail: omry.koren@biu.ac.il.
References
Author notes
These authors contributed equally to this study.
Requests for access to the study data can be e-mailed to the corresponding author: shouval@gmail.com.
The full-text version of this article contains a data supplement.