Key Points
Eight novel RHD variant alleles have been detected in the Finnish population following NGS of the whole RHD gene.
NGS is a powerful method for genotyping the highly polymorphic RHD gene.
Abstract
Fetal RHD screening for targeted routine antenatal anti-D prophylaxis has been implemented in many countries, including Finland, since the 2010s. Comprehensive knowledge of the RHD polymorphism in the population is essential for the performance and safety of the anti-D prophylaxis program. During the first 3 years of the national screening program in Finland, over 16 000 samples from RhD− women were screened for fetal RHD; among them, 79 samples (0.5%) containing a maternal variant allele were detected. Of the detected maternal variants, 35 cases remained inconclusive using the traditional genotyping methods and required further analysis by next-generation sequencing (NGS) of the whole RHD gene to uncover the variant allele. In addition to the 13 RHD variants that have been previously reported in different populations, 8 novel variants were also detected, indicating that there is more variation of RHD in the RhD− Finnish population than has been previously known. Three of the novel alleles were identified in multiple samples; thus, they are likely specific to the original Finnish population. National screening has thus provided new information about the diversity of RHD variants in the Finnish population. The results show that NGS is a powerful method for genotyping the highly polymorphic RHD gene compared with traditional methods that rely on the detection of specific nucleotides by polymerase chain reaction amplification.
Introduction
Immunization against the RhD antigen is the major cause of severe hemolytic disease of the fetus and newborn (HDFN).1 Thus, a comprehensive anti-D prophylaxis program is required to prevent immunizations. In addition to the postnatal prophylaxis implemented in the 1960s, routine antenatal anti-D prophylaxis is also now offered by many countries during the third trimester of pregnancy.2 The development of noninvasive fetal RHD genotyping has enabled the targeting of routine antenatal anti-D prophylaxis only to women carrying an RhD+ child, which prevents up to 40% of pregnant RhD− women carrying an RhD− fetus from receiving unnecessary prophylaxis.3
Finland implemented national fetal RHD screening for targeted anti-D prophylaxis for all RhD− pregnant women in February 2014. Fetal RHD screening is offered to all women who have been typed serologically as D−. Pregnant women with partial D variants and certain weak D variants, as well as those who have inconsistent phenotyping and genotyping results, are also considered RhD−, so that they are included in the prophylaxis program, due to the risk of immunization.
A maternal RHD variant can be suspected in the fetal RHD screening based on a low cycle-threshold value in the real-time polymerase chain reaction (PCR) assay, indicative of the high amount of maternal DNA present. The PCR-based genotyping methods primarily used for the identification of maternal variants rely on the detection of specific nucleotides by PCR amplification, so that only a limited number of known variants can be identified. Novel variants outside of the targeted regions therefore remain undetermined.4,5
The development of massively parallel high-throughput next-generation sequencing (NGS) has offered new possibilities for blood group genotyping.4,6-8 NGS can be efficiently applied for RHD genotyping, although the high level of homology between the RHD and RHCE genes is a challenge.4,9-13 The NGS approach can sometimes offer the advantage of detecting the cis∕trans linkage of polymorphisms, revealing heterozygous compound mutations, which cannot be identified in the conventional Sanger sequencing due to signal overlay in the electropherogram.
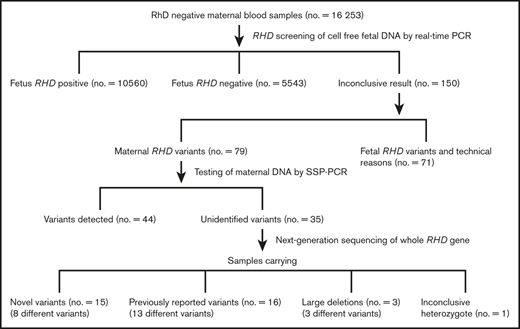
During the first 3 years of the national Finnish screening program, 16 253 samples of D− pregnant women were screened for fetal RHD. From this cohort, 79 maternal RHD variants were detected. Of the detected variants, 35 samples needed further scrutinizing because the sequence-specific primer PCR (SSP-PCR) method indicated the presence of a normal RHD gene although it did not correspond to the phenotype of the samples. In this study, these samples were genotyped by NGS for the complete RHD gene to identify the causative mutations for the negative or inconclusive RhD-phenotyping results.
Materials and methods
All 16 253 samples were from pregnant RhD− women who participated in the national screening program for HDFN in Finland between February 2014 and December 2016. The sample collection and processing was performed as described by Haimila et al.14
The study was approved by the Research Ethics Committee of Helsinki University Hospital Women’s clinic (271/13/03/03/2014) and by the National Institute for Health and Welfare (1365/5.05.01/2014).
RhD phenotyping
All samples were phenotyped for RhD using a solid-phase microplate system by Olympus (PK 7300 automate; Beckman Coulter). Two monoclonal anti-D reagents were used, both in 2 dilutions: the clone LDM3/ESD1 (Alba Bioscience Limited) and the clone TH-28/MS-26 (Bioscot; Millipore). If the premier method did not give a clear positive (n.b.: RhD+ samples were not included in this study) or negative result, an IH-1000 instrument (Bio-Rad) was also used to test a sample for RhD according to the manufacturer’s instructions. The anti-D reagent used in the IH-1000 instrument was LHM 59/20 (LDM3) in ABO/D reverse grouping gel cards (Bio-Rad).
All samples giving a clear negative result, or a weak positive reaction (≤2+) and a genotype not corresponding to the phenotype leaving the RhD result inconclusive, were designated as RhD−, meaning that women were included in the anti-D prophylaxis program to prevent possible anti-D immunization.
To estimate the amount of D antigen on red blood cells, an absorption-elution test was performed. For anti‐D adsorption, 700 μL of washed red blood cells (RBCs) were incubated for 1 hour at 37°C with an equal volume of anti-D (anti-D [Rh1], monoclonal, P3x61; Diagast). The cells were washed 5 times (Titriplex III 0.09 mol/L, disodium hydrogen phosphate dihydrate 0.026 mol/L, NaCl 0.148 mol/L, pH 6.8). Elution was effected by an acid-elution buffer, pH 2.8, and neutralized by an alkaline buffer, pH 10.4. The eluate and the last washed supernatant were used for the direct antiglobulin test against D+ (CcDEe) reagent RBCs. Serial dilutions of eluate in phosphate-buffered saline were prepared. Next, 60 μL of 4% D+ RBCs were added to each tube, incubated 15 minutes at 37°C, and centrifuged at 500g for 10 seconds. The agglutination was read macroscopically and the reciprocal of the highest eluate dilution that gave a 1+ agglutination reaction was interpreted as the titer.
RHD genotyping
All 16 253 samples were screened for fetal RHD as described by Haimila et al.14 In 79 samples, a maternal variant was suspected based on low cycle-threshold values, and maternal genomic DNA was extracted from the original sample using the QIAsymphony SP automated DNA-extraction method and the QIAamp DNA Blood Midi kit (Qiagen) according to the manufacturer’s instructions. The maternal DNA samples were then tested for RHD and RHCE using the SSP-PCR RBC-Ready Gene CDE kit and/or the weak D kit (inno-train Diagnostik) according to the manufacturer’s instructions. The RHD result still remained inconclusive in 35 cases and these samples were followed up with sequencing.
RHD zygosity testing by droplet digital PCR
All 35 maternal samples containing unidentified RHD variants were tested for RHD zygosity using the QX100 droplet digital PCR (ddPCR) platform and QuantaSoft v1.7 software (Bio-Rad Laboratories), as described in Sillence et al,15,16 to determine whether the women carried 1 (hemizygous Dd) or 2 copies (homozygous DD) of the RHD gene. Primers and probes used in ddPCR zygosity testing were adopted from Fan et al17 and Finning et al.18 The samples were tested for RHD exons 5 and 7, against the reference gene AGO1 on chromosome 1. The number of RHD copies per microliter for each exon was compared with the reference gene (AGO1) copy number. A sample giving a ratio of ∼0.5 was considered hemizygous and a sample giving a ratio of ∼1 was considered homozygous for the RHD gene.
RHD genotyping using NGS
In all 35 maternal samples containing unidentified RHD variants, the RHD gene was amplified in 6 overlapping amplicons using long-range PCR (LR-PCR) primers described in Tounsi et al.9 Primers from Hyland et al19 were also used to amplify the RHD gene from exon 2 to exon 7 in a 22 117-bp amplicon for 1 sample that failed to amplify amplicons 1, 2, and 3 of the 6 LR-PCR RHD amplicons. The sequencing library was prepared as in Tounsi et al.9 Amplicons were attached to ion-sphere particles using the Ion PGM Template OT2 400 kit (Life Technologies) according to the manufacturer’s instructions. Multiplexed sequencing was conducted using the Ion Xpress Barcode Adapters (Life Technologies) and a 316 chip (Life Technologies) on an Ion Torrent Personal Genome Machine (Life Technologies) as per manufacturer guidelines.
NGS data analysis
Quality control of the sequencing reads was done using Babraham Bioinformatics FastQC software20 quality check. Data analysis of the sequencing FASTQ data were performed using CLC Genomics Workbench software (version 11.0.1; Qiagen). To map the reads to the reference, the RHCE gene and other genes on chromosome 1 assembly were masked to prevent the reads from scattering. The mapping was performed using the default parameters with NC_000001.11 (Homo sapiens chromosome 1, GRCh38.p13 Primary Assembly) as reference.21,22
In the basic variant detection, a minimum coverage of 30, a minimum count of 5, and a minimum frequency of 80 were used for the samples considered hemizygous based on the ddPCR results. For the sample containing 2 copies of the RHD gene, a minimum frequency of 40 was used. The single-nucleotide polymorphisms (SNPs) detected that predicted amino acid or splice site changes were compared with known RHD variants on RhesusBase,23 Erythrogene,24 and the Blood Group Antigen FactsBook25 to determine the variant RHD allele.
Sanger sequencing
The PCR primers used in the verification of the novel 829G>A mutation were 5′-AGTAGTGAGCTGGCCCATCA-3′ and 5′-AGCAGAGGAGGTTAGTTGTCT-3′. The latter was used as a sequencing reverse primer as well, whereas 5′-TCTAGTTTCTTACCGGCAGGT-3′ was used as a sequencing forward primer. PCR was set up using AmpliTaq DNA polymerase according to the manufacturer’s instructions (Thermo Fisher Scientific), using both primers at 0.2-μM concentration. The sequencing of the PCR products was performed by the DNA Sequencing and Genomics Unit of the Institute of Biotechnology, Helsinki Institute of Life Science (HiLIFE), University of Helsinki.
Results
RHD zygosity
ddPCR results indicated that all samples except for 1 were hemizygous for RHD (Table 1), showing ratios of ∼0.5 for both RHD5/AGO1 and RHD7/AGO1. However, 1 sample showed both RHD5/AGO1 and RHD7/AGO1 ratios of 1, which indicated that it carried 2 copies of the RHD gene (homozygous).
Results of the samples (n = 35) genotyped for the RHD gene using NGS
 |
 |
Gray indicates D− alleles; green, DEL alleles; blue, weak D alleles; orange, partial D alleles; yellow, novel variant alleles; and red, inconclusive results.
Del, deletion; Ins, insertion; NA, not applicable; Neg, negative; NR, not reported; pos/Pos, positive; SNV, single-nucleotide variation.
International Society of Blood Transfusion (ISBT) designation for alleles.
Linked haplotype reported in the RhesusBase database.
To assign the most possible DCE haplotype, RhCcEe genotyping results, RHD zygosity results, and RHD allele determined by NGS were all considered.
Allele frequency was calculated based on the total number of RhD− females (16 253).
Routine RhD phenotyping was negative, but antigen titer was 1 after elution.
RHD*11 occurs in 2 haplotypes with distinct phenotypes. In the DCe haplotype, the phenotype is borderline weak D/DEL, whereas in the Dce haplotype, the phenotype is weak D detected in routine tests.
Samples failed amplicon 6 in LR-PCR, thus incomplete data.
Sample failed amplicons 1, 2, and 3 in LR-PCR.
This sample had RHD5/AGO1 and RHD7/AGO1 ratios of ∼1 from ddPCR and was considered homozygous for the RHD gene. The NGS data indicated that the sample is a compound heterozygote with 2 variant RHD alleles.
Next-generation sequencing
NGS of the whole RHD gene revealed 21 different variant RHD alleles in 31 of 35 samples (Table 1). In total, 13 previously described RHD variants were identified in 16 samples. These variants include 3 null alleles, 3 DEL alleles, 3 weak D alleles, 3 partial weak D alleles, and 1 allele previously reported by Karnot et al31 for which there is no designation by International Society of Blood Transfusion (ISBT) as found in RhesusBase.23 Mutations that encode the variant alleles and the predicted amino acid changes are listed in Table 1.
In total, 8 different novel alleles were identified in 15 samples, and these are encoded by 4 novel missense mutations, 3 frame-shift mutations caused by deletions, and 1 nonsense mutation (Table 1). Reference sequences of the novel RHD variant alleles were submitted to GenBank and were each assigned a unique accession number, and the allele frequencies were calculated based on the number of RhD− women (16 253) included in the Finnish national program to prevent HDFN (Table 1). The novel SNP 829G>A in exon 6 that predicts amino acid change Gly277Arg was confirmed using Sanger sequencing because the same nucleotide change in the adjacent base, 830G>A, has been reported to cause the RHD*01W.12 (RHD*weak D type 12) allele.
Two mutations found in exon 9 (1154G>C, 1163T>G) predicting (Gly385Ala, Leu388Arg) were detected in 1 sample. The mutation 1154G>C encodes RHD*01W.2 but the allele was considered a novel variant due to the additional mutation detected. Similarly, 2 mutations (8C>G, 49delG) in exon 1 that predict Ser3Cys and a frame-shift Ala17fs were detected in 1 sample. The mutation 8C>G encodes RHD*01W.3 but the allele was considered a novel variant due to the presence of the frame-shift SNP.
In 3 samples, LR-PCR reactions of 1 or multiple RHD amplicons failed presumably due to a hybrid gene or a deletion preventing the annealing of the RHD-specific LR-PCR primers (Table 1). In 2 of these samples, the failed amplicon was amplicon 6, which includes the area between part of intron 8 to exon 10 of the RHD gene. In the third sample, the LR-PCR reactions of RHD amplicons 1, 2, and 3 failed, which cover the region between RHD exons 1 and 5. The LR-PCR was repeated for this sample using primers adopted from Hyland et al19 to amplify the RHD gene from exon 2 to exon 7. The results showed a complete deletion of exon 3 (336-486del) with a partial deletion of intron 2, in addition to mutations detected in exons 4, 5, and 6 (602C>G, 667T>G, 819G>A) that predict amino acid changes (Thr201Arg, Phe223Val, silent), respectively.
In 1 sample, which showed a homozygous RHD gene using ddPCR, 4 mutations (48G>C, 602C>G, 667T>G, and 819G>A) were detected in exons 1, 4, 5, and 6, respectively. These mutations predict the amino acid changes Trp16Cys, Thr201Arg, Phe223Val, and a silent mutation, respectively. These changes encode RHD*09.04, meaning that the other allele is a wild-type RHD allele RHD*01. However, the presence of a wild-type RHD allele that produces a normal RhD protein does not agree with the weak D reactivity in serology.
Discussion
In Finland, genotyping is not performed unless there is an indication of an RHD variant. Therefore, variants that cause no or very low expression of the D antigen are not detected in the serological tests. Thus, the true variation of RHD among the RhD− population in Finland has not been known. This study, encompassing a cohort of over 16 000 RhD− pregnant women screened for fetal RHD during the first 3 years of the national program to prevent HDFN, is the first comprehensive study of RHD genetics in the Finnish population. NGS genotyping of the cases that remained inconclusive using SSP-PCR revealed 21 different RHD variants, of which 8 were novel alleles likely specific to the original Finnish population. All novel alleles were found in women who had been born in Finland and who had typical Finnish names. The SNPs 829G>A (rs1456633324), 421delG (rs755478959), and 422T>A (rs759038817) are listed in genome-aggregation database32 and have been detected in European (Finnish) samples, with the following frequencies: 0.00001794, 0.000008886, and 0.000008886, respectively.
RHD variation in the Finnish population
The frequency of RHD variants varies between populations and depends on the structure and amount of admixture in the population. The frequency of maternal variants detected in this study was 0.5%, which is similar to the frequencies detected in Denmark33 and The Netherlands34 with national programs. The frequency of RHD+ variants among RhD− blood donors is 0.47% in Switzerland35 and 0.21% in Germany.26 The known variants detected in this study have been previously reported in different populations and thus reflect the effect of increasing immigration and admixture in the Finnish population. However, the variety and frequency of novel variants was surprisingly high and reflects the lack of knowledge of the diversity of RHD variants among the RhD− population in Finland. For example, Crottet et al35 discovered 4 novel RHD alleles among 25 370 RhD− blood donors in a large national screening in Switzerland; de Paula Vendrame et al36 found 6 novel alleles in a population of 1920 Brazilian RhD− blood donors.
Novel RHD variants
In this study, 8 novel mutations of RHD were detected that likely constitute novel null, weak D, and DEL variants, although the phenotypes could not be confirmed because fresh samples of the variant carriers were not available for D antigen–quantifying tests. The novel 421delG, 784delC, and 519C>G mutations are expected to cause null phenotypes, which would be consistent with the observed D− results in the RhD phenotyping of the samples. In addition, 3 novel missense mutations were detected. The SNPs 829G>A, 782C>T, and 1016G>C all cause amino acid changes in the transmembrane part of the RhD protein and can thus be expected to cause a weak D or DEL phenotype. All 3 samples that were predicted to have the amino acid change Pro261Leu (782 C>T) or Gly339Ala (1016 G>C) showed weak reactions in RhD phenotyping, which is consistent with the weak D phenotype caused by missense mutations in the transmembrane parts of the protein.
On the other hand, all 5 samples with the 829G>A mutation predicting amino acid change Gly277Arg showed negative reactions in the phenotyping, which could indicate a DEL phenotype because missense mutations should not cause a null phenotype. The mutation 830G>A, that is, the adjacent base, causes the amino acid change Gly277Glu and the RHD*01W.12 variant allele, which has been associated with extremely low antigen density.37 In both the 829G>A and 830G>A mutations, a nonpolar amino acid (glycine) is changed either to a positively charged (arginine; 829 G>A) or negatively charged (glutamic acid; 830 G>A) amino acid, so the novel Gly277Arg change observed in the samples could have similar consequences to the RhD polypeptide as the Gly277Glu change in RHD*01W.12. However, the possible DEL phenotype of 829G>A causing Gly277Arg would need to be confirmed with D antigen–quantifying tests.
Two novel mutations were discovered on a backbone of RHD*01W.2 and RHD*01W.3 alleles. These samples harbored not only the mutations responsible for the mentioned alleles, but also additional mutations that may explain the negative serological results of the samples. The deletion 49delG leads to a frame shift and thus probably a nonfunctional RhD protein. The missense mutation 1163T>G predicts the amino acid change Leu388Arg, which is located in the transmembrane region of the RhD protein. Whether the phenotype in the sample is DEL could only be confirmed with antigen-quantifying tests. The frequency of weak D types 1, 2, and 3 was ∼0.2% in this study period (including also RhD+ samples) and the frequency of the novel mutations on the backbone of RHD*01W.2 and RHD*01W.3 alleles is 0.8% (2 of 242 weak D type 1, 2, and 3 samples). Relying solely on single SNP-genotyping assays designed to detect weak D mutations, a sample could be misassigned as weak D type 2 or 3 rather than D−. However, additional mutations can cause a risk of immunization. Novel alleles are often population specific38 and, thus, commercial genotyping methods based on the detection of known mutations cannot unveil them. LR-PCR NGS genotyping can improve the safety of blood transfusion practices by revealing novel mutations. When serologically weak D phenotypes are detected, laboratories should complete RhD testing by determining RHD genotypes because weak D type 1, 2, 3, 4.0, and 4.1 individuals can be safely transfused with D+ blood without being at risk for forming alloanti-D.39,40 But, if the phenotype and genotype do not correspond, the sample should be sequenced or the person managed as RhD−.
Known RHD variants
Among the previously reported RHD variants identified in this study, the Rh-phenotyping results of the samples corresponded to the reported phenotypes of the variant alleles in all but 3 samples.
Weak D type 1, 2, and 3 are the most common RHD variants in White populations.29 They are usually detected in the serological RhD typing and identified using the SSP-PCR method. Because women carrying weak D type 1, 2, or 3 are not considered to be at risk of immunization and do not need anti-D prophylaxis, they can be excluded from the fetal RHD screening. The detection of RHD*01W.2 and RHD*01W.3 alleles in 3 samples was thus surprising. The 3 samples identified as RHD*01W.2 or RHD*01W.3 had given negative or very weak reactions in the serological RhD typing and for that reason were included in the screening program.
The suppressing effect of C in the in trans haplotype, the Ceppellini effect,41 can cause the reduction of D-antigen density and lead to a DEL phenotype in weak D samples.42,43 In the 3 samples identified as RHD*01W.2 and RHD*01W.3 in this study, no additional mutations were found, but they all have the Ce haplotype in trans to the weak D allele, which could explain the decrease in D-antigen expression in these samples. These results are consistent with previous observations of a very low antigen density in weak D samples that have C in trans.37,44,45 An antigen adsorption-elution test to quantify D-antigen expression could only be performed for 1 sample of the 3, for which a fresh blood sample was available. The adsorption-elution test indicated an antigen titer of 1, which denotes a very low antigen density and a DEL phenotype.
In the samples in which a previously reported variant was identified, the probable variant haplotypes were consistent with the reported variant haplotypes in all but 1 sample. The RHD*15 allele is usually associated with antigen E and observed in the DcE variant haplotype.29 However, Ye et al30 observed samples that were identified as RHD*15 also in the ee (Cce) phenotype. In 1 of the 2 samples identified as RHD*15 in this study, the most likely variant haplotype is DCe (given RhCcEe-genotyping results, RHD zygosity results, and the RHD allele determined from NGS), which supports the observation of RHD*15 also being associated with e.
Advantage of NGS
The current methods used in RHD genotyping, such as SSP-PCR and real-time PCR, are insufficient to identify many variants because they require the causative mutations to be known. During the first 3 years of the national fetal RHD screening program in Finland, almost one-half of the detected maternal variants could not be identified (35 of 79). The SSP-PCR results, however, indicated inconsistency between phenotype and genotype and thus, a variant allele.
The results of the use of NGS in blood group genotyping to overcome limitations in other genotyping platforms have been promising.9,11,12,46 Exome sequencing utilizing NGS or conventional sequencing (Sanger) could have been used to identify variants; however, exome sequencing may not have been sufficient to detect intronic variations such as intronic insertion associated with RHD partial alleles.12 Intronic mutations detected in this study align with those in Tounsi et al9 for R2 and R1/R0/RZ haplotypes. NGS allows the use of the LR-PCR approach, enabling the amplification of the RHD gene (<60 kb) in only 6 overlapping amplicons about 10 kb in size. On the other hand, Sanger sequencing cannot sequence DNA fragments larger than 2 kb.47 Therefore, complete sequencing of the RHD gene using NGS was the preferred method, which provided a complete sequence of the gene in most samples, with high coverage that allowed accurate variant calling. The advantage of NGS over Sanger sequencing is that it has the ability to sequence the genes for multiple blood group antigens simultaneously (and not just RHD) and hence can be applied in routine workflows in reference laboratories looking at samples with complex variants,48 as in this study.
NGS of the whole RHD gene in this study was efficient in detecting the exonic and intronic changes (Table 1 and data not shown) causing the variants of RHD, except for the hemizygous hybrid RHD-CE-D genes and/or large deletions. Hybrid genes and large deletions can cause the PCR amplicons to fail if they affect the RHD-specific LR-PCR primer-binding sites. In 2 samples, LR-PCR amplicon 6 covering the area from intron 8 to exon 10 of the RHD gene failed. In these cases of failed amplicons, other methods, like single-molecule sequencing, would be needed in order to identify the exact location and extent of the changes. In the sample in which amplicons 1, 2, and 3 covering the region between exons 1 and 5 of the RHD gene had failed, using a different approach with primers surrounding the failed area revealed a large deletion covering part of intron 2 and all of exon 3, and an additional mutation in exon 4 in the failed region. In the future, oligonucleotide bait enrichment may allow for the sequencing of the entire RHD gene (or hybrid gene) directly from genomic DNA without the need for PCR amplification.49
In 1 RHD homozygous sample, the variant alleles could not be determined. Four heterozygous SNPs were detected that encode RHD*09.04, indicating that the other RHD allele is a wild-type RHD allele. This does not agree with weak D serological results. As serological phenotyping was weak D, it indicates that the variant carrier is a compound heterozygote for 2 different variant alleles. The other variant RHD allele is possibly a variant RHD allele that harbors a deletion or a conversion that could not be detected due to the presence of an intact copy of the RHD gene (RHD*09.04).
This was the first comprehensive study performed in the Finnish RhD− population and revealed previously unknown allelic diversity behind the negative RhD phenotype. NGS genotyping of the entire RHD gene proved to be cost-efficient in uncovering the mutations causing the RHD variants that could not be identified using the traditional SSP-PCR technique and revealed 8 novel variants that we believe are linked to the original Finnish population. The identification of RHD variants causing inconclusive typing results is essential in order to prevent immunizations in pregnancies and transfusions.
The novel RHD variants reported in this article have been deposited in the GenBank database (accession numbers MN365996, MN365995, MN366000, MN365998, MN366001, MN365997, MN365999, and MN366002).
Acknowledgments
This work was supported by King Abdulaziz University (Jeddah, Saudi Arabia).
Authorship
Contribution: S.S. and K.H. designed the study; S.M.T. and K.H. collected the data of the samples; S.M.T., W.A.T., and M.K. performed experiments; S.M.T., W.A.T., T.E.M., and K.H. analyzed the data and wrote the manuscript; T.E.M., N.D.A., and K.H. supervised the study; and all authors reviewed, edited, and approved the manuscript.
Conflict-of-interest disclosure: N.D.A. is a consultant for YourGene Health plc. The remaining authors declare no competing financial interests.
Correspondence: Katri Haimila, Finnish Red Cross Blood Service, Kivihaantie 7, FI-00310 Helsinki, Finland; e-mail: katri.haimila@bloodservice.fi.
References
Author notes
S.M.T. and W.A.T. contributed equally to this study.