Key Points
HLA-A*24 and HLA-A*32 inhibit KIR3DL1+ NK cells and contribute to NK education.
The combination of donor KIR3DL1 and HLA-A*24 is associated with higher risk of AML relapse following allogeneic HCT.
Abstract
HLA-B allotypes exhibiting the Bw4 epitope trigger variable inhibitory signaling of KIR3DL1 receptor types, where strong inhibitory HLA-B and KIR3DL1 allele combinations are associated with increased risk for relapse of acute myelogenous leukemia (AML) following allogeneic hematopoietic cell transplantation (HCT). Several HLA-A allotypes also exhibit the Bw4 epitope. Studies with natural killer (NK) cell clones have demonstrated NK inhibition via KIR3DL1 by HLA-A Bw4+ allotypes, but did not delineate strengths of inhibition or hierarchies of NK education. Using primary NK cells from healthy donors, we demonstrate that HLA-A*23, HLA-A*24, and HLA-A*32 proteins are expressed at different densities and exhibit different capacities to educate and inhibit KIR3DL1-expressing NK cells in vitro. Among the HLA-A Bw4+ allotypes, HLA-A*24 and HLA-A*32 demonstrate the strongest inhibitory capacity. To determine if HLA-A allotypes with strong inhibitory capacity have similar negative impact in allogeneic HCT as HLA-B Bw4+ allotypes, we performed a retrospective analysis of 1729 patients with AML who received an allogeneic HCT from a 9/10 or 10/10 HLA allele-matched unrelated donor. Examination of the donor-recipient pairs whose Bw4 epitope was exclusively contributed from HLA-A*24 and A*32 allotypes revealed that patients with HLA-A*24 who received an allograft from a KIR3DL1+ donor experienced a higher risk of disease relapse (hazard ratio, 1.65; 95% confidence interval, 1.17-2.32; P = .004) when compared with patients without a Bw4 epitope. These findings indicate that despite weak affinity interactions with KIR3DL1, common HLA-A allotypes with the Bw4 epitope can interact with KIR3DL1+ donor NK cells with clinically meaningful impact and provide additional insight to donor NK alloreactivity in HLA-matched HCT.
Introduction
Natural killer (NK) cells eliminate virally infected and transformed cells through the engagement of activating receptors in the absence of inhibitory ligands. Inhibitory NK cell receptors are central to self-tolerance via their engagement with class I HLA on target cells. These same receptors and their HLA class I ligands are essential to the acquisition of functional competence in a process referred to as NK cell education. Chief among the inhibitory receptors are the killer immunoglobulin-like receptors (KIR), which are encoded by the highly polymorphic KIR gene family.1 NK cells that express inhibitory KIR for self-HLA are educated and more responsive than their uneducated counterparts,2,3 but are also more sensitive to inhibition.
Acute myelogenous leukemia (AML) is sensitive to NK cell killing,4,5 suggesting that NK alloreactivity plays an important role in defining the graft-versus-leukemia effect after allogeneic hematopoietic cell transplantation (HCT). Selection of allogeneic stem cell donors with favorable NK immunogenetic conditions may skew NK cell activity in vivo toward tumor lysis, improving outcomes in AML patients undergoing HCT. Early studies highlighted that donor inhibitory KIR and recipient HLA class I mismatch minimizes NK cell inhibition, resulting in greater NK cell reactivity and lower AML relapse.6-9 Among the assortment of KIR receptors and their ligands, the inhibitory KIR3DL1 receptor and HLA-B ligands bearing the Bw4 epitope are strongly associated with outcomes in viral infection and malignancy, including allogeneic HCT for AML.7,10-12 The highly polymorphicKIR3DL1 alleles encode receptor variants with different cell surface densities ranging from high (KIR3DL1*001, *002, *015) to low (KIR3DL1*005 and *007), with still one other common variant retained in the cell (KIR3DL1*004).13,14 KIR3DL1 is known to bind the Bw4 epitope at amino acids 77 to 83 on the α1-helix of the HLA molecule, where the amino acid at position 80 critically determines binding to KIR3DL1.15-18 Allotypes with the Bw4 epitope have either an isoleucine (I80) or threonine (T80) at this position, whereas allotypes with the Bw6 epitope exhibit an asparagine residue at position 80, precluding KIR3DL1 binding.18,19 Specific combinations of KIR3DL1 expression subtype and the HLA-B Bw4 dimorphism (I80/T80) display an array of binding strength, leading to strong or weak NK cell inhibition and education.11,16,17 In AML patients undergoing allogenic HCT from a 9/10 or 10/10 HLA-matched donor, donor-recipient KIR3DL1/HLA-B Bw4 subtype combinations with weak interaction are associated with significantly lower relapse and higher survival when compared with donor-recipient pairs with strong inhibition KIR3DL1/HLA-B Bw4 combinations.11
Not sufficiently considered in these genetic association studies are the HLA-A allotypes that exhibit the Bw4 epitope, specifically the HLA-A*23, HLA-A*24, HLA-A*25, and HLA-A*32 molecules, which express the Bw4-I80 subtype and bind KIR3DL1 with highly variable specificities.16-18,20-23 Prior binding studies of Bw4+ HLA-A proteins used Fc recombinant proteins, tetramers, and NK cell clones to demonstrate the ability of HLA-A Bw4+ molecules to inhibit NK cells without clear implication for function in vivo.16-18,20-23 Here, we sought to determine the inhibitory and educating capacities of the HLA-A Bw4+ allotypes, using primary KIR3DL1+ NK cells from healthy human donors. Similar to findings with NK clones, primary KIR3DL1+ NK cells are inhibited and educated by HLA-A*24 and HLA-A*32. The relatively high population frequency of HLA-A*24 permitted examination of its relevance to transplant outcome in AML patients receiving allogeneic HCT from HLA-compatible donors. We find that similar to high inhibitory KIR3DL1/HLA-B pairs, the combination of donor KIR3DL1 and HLA-A*24 is associated with increased relapse in AML patients undergoing HCT. Together, these findings indicate that HLA-A alleles, in particular HLA-A*24, should be considered when using immunogenetics to predict NK cell alloreactivity in donor selection for allogeneic HCT.
Methods
Clinical samples and healthy donor peripheral blood mononuclear cells
We studied 1729 patients with AML who underwent allogeneic HCT and received an allograft from a 9/10 or 10/10 HLA allele-matched unrelated donor. All 9/10 matched donor/recipient pairs were matched for HLA-A-Bw4+ ligand. Only donor-patient pairs for whom HLA typing and donor DNA were available were selected for analysis. The Center for International Blood and Marrow Transplant Research provided clinical data, HLA genotyping, and genomic DNA for KIR genotyping. Studies were executed in compliance with federal regulations pertaining to the protection of human research participants and were approved by the Center for International Blood and Marrow Transplant Research Institutional Review Board.
Following approval from the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board, peripheral blood mononuclear cells (PBMCs) from anonymous healthy donors were isolated from buffy coats obtained from the New York Blood Center (New York, NY) using Ficoll centrifugation. Fetal bovine serum with 10% dimethyl sulfoxide was used to cryopreserve isolated PBMCs. Informed consent for research was obtained from all individuals.
KIR gene typing, KIR3DL1 allele typing, and HLA class I genotyping
KIR gene typing and KIR3DL1 subtyping for patients and healthy donors were performed as previously described.17,24-27 KIR3DL1high, KIR3DL1low, KIR3DS1, or KIR3DL1null (KIR3DL1*004) subtypes were used to classify the KIR3DL1 alleles, as described in previous studies.11,17 HLA-A, HLA-B, and HLA-C allele typing for healthy donors was completed by Histogenetics (Ossining, NY) and alleles encoding HLA-Bw4-I80, HLA-Bw4-T80, and HLA-Bw6 subtypes were assigned using the IPD-HLA database, version 3.34.0.
Assessment of NK cell activation by flow cytometry
CD107a mobilization and interferon-γ (IFN-γ) production were measured to determine NK cell activation. Frozen PBMCs were thawed and rested overnight in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 200 U/mL IL-2 (Proleukin, Prometheus Laboratories), and incubated at 37°C with 5% CO2. In V-bottom 96-well plates, PBMCs (2.5 × 105 cells per well) were incubated with target cells at a 1:5 ratio in the presence of anti-CD107a antibody (BD Biosciences, San Jose, CA). As target cells, Epstein-Barr virus-transformed B-lymphocyte cell lines (BLCLs; kindly provided and generated by Richard O’ Reilly of MSKCC or derived from Centre d’Etude Polymorphisme Humaine [http://www.cephb.fr/en/familles_CEPH.php]) (Table 1) and K562 cells (ATCC, Manassas, VA) were used and cultured in RPMI complete media. To measure IFN-γ production, BD GolgiStop (BD Biosciences) was added to the cells at a 1:1500 dilution after 1 hour of coculture. For inhibition experiments, PBMCs were cultured with or without 10 μg/mL anti-KIR3DL1 (DX9 clone) antibody for 15 minutes at room temperature and NK cells were activated for antibody-dependent cellular cytotoxicity (ADCC) by the addition of 0.1 μg/mL rituximab before coculture with BLCL target cells. After 5 hours of coculture with the target, cells were stained using the antibodies shown in supplemental Table 1. In all experiments, NK cells exclusively expressing KIR3DL1 were analyzed, excluding cells coexpressing other inhibitory receptors that could contribute to NK education (KIR2DL1, KIR2DL2, KIR2DL3, and NKG2A) and cells coexpressing activating receptors that could enhance NK effector function (KIR2DS1, KIR2DS2, and NKG2C). Inhibition of NK cells via KIR3DL1 by HLA-A Bw4+ allotypes was calculated as the percent decrease of activation in culture with the target cells compared with culture with the same cells in the presence of the KIR3DL1-blocking clone DX9. Baseline activation, assessed as the frequency of NK cells expressing CD107a, and percent decrease upon inhibition were calculated for each donor-target pair, normalizing for donor variation: 100 × [1 − (NK + Target)/(NK + Target + DX9)].
BLCL HLA class I genotype
| BLCL name . | HLA-A . | HLA-B . | HLA-C . |
|---|---|---|---|
| B*38/B*52(I80) | A*11:01/A*26:01 | B*38:01/B*52:01 | C*12:02/C*12:03 |
| Bw6 | A*02:01/A*11:01 | B*08:01/B*55:01 | C*07:01/C*03:03 |
| A*24 | A*24:02/A*24:02 | B*40:06/B*40:06 | C*12:02/C*15:07 |
| A*32 | A*32:01/A*01:01 | B*35:03/B*40:02 | C*04:01/C*15:02 |
| BLCL name . | HLA-A . | HLA-B . | HLA-C . |
|---|---|---|---|
| B*38/B*52(I80) | A*11:01/A*26:01 | B*38:01/B*52:01 | C*12:02/C*12:03 |
| Bw6 | A*02:01/A*11:01 | B*08:01/B*55:01 | C*07:01/C*03:03 |
| A*24 | A*24:02/A*24:02 | B*40:06/B*40:06 | C*12:02/C*15:07 |
| A*32 | A*32:01/A*01:01 | B*35:03/B*40:02 | C*04:01/C*15:02 |
HLA alleles with a Bw4 epitope are indicated in bold. HLA-B*38 and HLA-B*52 have an isoleucine at amino acid position 80.
HLA-A Bw4+ allotype surface staining
PBMCs of healthy donors with HLA-A genotypes inclusive of HLA-A*24, HLA-A*32, HLA-A*23, or HLA-A*25 and with HLA-B genotypes encoding molecules exhibiting only the HLA-Bw6 epitope were thawed and rested overnight in RPMI-1640 complete medium. For HLA-A Bw4+ allotype surface staining, 10 μL of fluorescein isothiocyanate-conjugated anti-HLA-Bw4 antibody (One Lambda) was used. PBMCs were also stained with DAPI (viability marker), anti-CD3, anti-CD56, and anti-CD19 antibodies (supplemental Table 1) to evaluate the surface expression of the HLA-A Bw4+ allotype on NK, T, and B cells. Flow cytometry data were analyzed using FlowJo software (v10.6.1, BD, Franklin Lakes, NJ).
Statistical analysis
Cox proportional hazards regression models were used for time-to-event post-HCT outcomes for relapse and death. Cumulative incidence estimates were used to calculate the probabilities of overall relapse. Multivariate analysis was performed without adjustments for multiple comparisons with the following covariates (Table 2): year of transplant, total body irradiation, age of patient and donor, conditioning regimen, T-cell depletion, graft type, disease status, cytomegalovirus serostatus, sex match, HLA-match, and donor KIR2DS1/HLA-C1. One-way analysis of variance (ANOVA) and χ2 tests were used to assess frequencies of clinically relevant covariates in the population grouped by recipient Bw4 content and donor KIR3DL1 allele group.
Patient and donor characteristics
| . | Overall . | Bw6/Bw6/non-A*32/*24 . | Bw6/Bw6/A*32 donor KIR3DL1pos . | Bw6/Bw6/A*24 donor KIR3DL1pos . | Bw6/Bw6/A*24 donor KIR3DL1neg . | |
|---|---|---|---|---|---|---|
| N | 604 | 451 | 25 | 110 | 18 | |
| GVHD prophylaxis, n (%) | ||||||
| CsA-based | 215 (35.6) | 162 (35.9) | 11 (44.0) | 38 (34.5) | 4 (22.2) | |
| Other | 55 (9.1) | 43 (9.5) | 1 (4.0) | 10 (9.1) | 1 (5.6) | |
| Tac-based | 334 (55.3) | 246 (54.5) | 13 (52.0) | 62 (56.4) | 13 (72.2) | |
| TBI-based conditioning, n (%) | ||||||
| Unknown | 2 (0.3) | 2 (0.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| No | 309 (51.2) | 234 (51.9) | 15 (60.0) | 49 (44.5) | 11 (61.1) | |
| Yes | 293 (48.5) | 215 (47.7) | 10 (40.0) | 61 (55.5) | 7 (38.9) | |
| Recipient age, mean (SD), y | 40.82 (17.69) | 40.68 (17.78) | 43.09 (18.58) | 40.75 (17.74) | 41.66 (14.79) | |
| Donor age, mean (SD), y | 32.68 (13.24) | 32.40 (12.84) | 37.11 (12.74) | 33.00 (14.56) | 31.35 (15.19) | |
| Recipient CMV serostatus, n (%) | ||||||
| Unknown | 8 (1.3) | 5 (1.1) | 0 (0.0) | 3 (2.7) | 0 (0.0) | |
| Negative | 223 (36.9) | 174 (38.6) | 7 (28.0) | 34 (30.9) | 8 (44.4) | |
| Positive | 373 (61.8) | 272 (60.3) | 18 (72.0) | 73 (66.4) | 10 (55.6) | |
| Donor CMV serostatus (%) | ||||||
| Unknown | 45 (7.5) | 31 (6.9) | 2 (8.0) | 12 (10.9) | 0 (0.0) | |
| Negative | 399 (66.1) | 303 (67.2) | 18 (72.0) | 66 (60.0) | 12 (66.7) | |
| Positive | 160 (26.5) | 117 (25.9) | 5 (20.0) | 32 (29.1) | 6 (33.3) | |
| Graft source, n (%) | ||||||
| BM | 287 (47.5) | 211 (46.8) | 10 (40.0) | 57 (51.8) | 9 (50.0) | |
| PBSC | 309 (51.2) | 235 (52.1) | 14 (56.0) | 51 (46.4) | 9 (50.0) | |
| BM, PBSC | 8 (1.3) | 5 (1.1) | 1 (4.0) | 2 (1.8) | 0 (0.0) | |
| Conditioning intensity, n (%) | ||||||
| Unknown | 23 (3.8) | 19 (4.2) | 1 (4.0) | 3 (2.7) | 0 (0.0) | |
| Nonmyeloablative | 19 (3.3) | 15 (3.5) | 1 (4.2) | 3 (2.8) | 0 (0.0) | |
| Reduced intensity | 70 (12.0) | 52 (12.0) | 3 (12.5) | 12 (11.2) | 3 (16.7) | |
| Myeloablative | 492 (84.5) | 365 (84.3) | 20 (83.3) | 92 (86.0) | 15 (83.3) | |
| T-cell depletion, n (%) | ||||||
| Yes | 198 (33.1) | 155 (34.6) | 7 (29.2) | 30 (27.5) | 6 (33.3) | |
| No | 401 (66.4) | 293 (65.0) | 17 (68.0) | 79 (71.8) | 12 (66.7) | |
| Unknown | 5 (0.8) | 3 (0.6) | 1 (4.0) | 1 (0.9) | 0 (0.0) | |
| HLA match, n (%) | ||||||
| HLA 10/10 matched | 384 (63.6) | 293 (65.0) | 15 (60.0) | 62 (56.4) | 14 (77.8) | |
| HLA 9/10 matched | 220 (36.4) | 158 (35.0) | 10 (40.0) | 48 (43.6) | 4 (22.2) | |
| Year of transplant, n (%) | ||||||
| 2005-2014 | 363 (60.1) | 274 (60.8) | 13 (52.0) | 63 (57.3) | 13 (72.2) | |
| 1989-2005 | 241 (39.9) | 180 (39.2) | 12 (48.0) | 47 (42.7) | 5 (27.8) | |
| Disease status at HCT, n (%) | ||||||
| First complete remission | 247 (40.9) | 179 (39.7) | 10 (40.0) | 49 (44.5) | 9 (50.0) | |
| Second or later complete remission | 156 (25.8) | 118 (26.2) | 5 (20.0) | 25 (22.7) | 8 (44.4) | |
| No remission | 160 (26.5) | 122 (27.1) | 8 (32.0) | 29 (26.4) | 1 (5.6) | |
| Other/unknown | 41 (6.8) | 32 (7.1) | 2 (8.0) | 7 (6.4) | 0 (0.0) | |
| KIR2DS1 HLA-C1/C2 donor, n (%) | ||||||
| KIR2DS1neg/C1/x | 395 (65.4) | 300 (66.5) | 21 (84.0) | 73 (66.4) | 1 (5.6) | |
| KIR2DS1pos/C1/x | 185 (30.6) | 135 (29.9) | 4 (16.0) | 29 (26.4) | 17 (94.4) | |
| C2/C2 | 24 (4.0) | 16 (3.6) | 0 (0.0) | 8 (7.2) | 0 (0.0) | |
| . | Overall . | Bw6/Bw6/non-A*32/*24 . | Bw6/Bw6/A*32 donor KIR3DL1pos . | Bw6/Bw6/A*24 donor KIR3DL1pos . | Bw6/Bw6/A*24 donor KIR3DL1neg . | |
|---|---|---|---|---|---|---|
| N | 604 | 451 | 25 | 110 | 18 | |
| GVHD prophylaxis, n (%) | ||||||
| CsA-based | 215 (35.6) | 162 (35.9) | 11 (44.0) | 38 (34.5) | 4 (22.2) | |
| Other | 55 (9.1) | 43 (9.5) | 1 (4.0) | 10 (9.1) | 1 (5.6) | |
| Tac-based | 334 (55.3) | 246 (54.5) | 13 (52.0) | 62 (56.4) | 13 (72.2) | |
| TBI-based conditioning, n (%) | ||||||
| Unknown | 2 (0.3) | 2 (0.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| No | 309 (51.2) | 234 (51.9) | 15 (60.0) | 49 (44.5) | 11 (61.1) | |
| Yes | 293 (48.5) | 215 (47.7) | 10 (40.0) | 61 (55.5) | 7 (38.9) | |
| Recipient age, mean (SD), y | 40.82 (17.69) | 40.68 (17.78) | 43.09 (18.58) | 40.75 (17.74) | 41.66 (14.79) | |
| Donor age, mean (SD), y | 32.68 (13.24) | 32.40 (12.84) | 37.11 (12.74) | 33.00 (14.56) | 31.35 (15.19) | |
| Recipient CMV serostatus, n (%) | ||||||
| Unknown | 8 (1.3) | 5 (1.1) | 0 (0.0) | 3 (2.7) | 0 (0.0) | |
| Negative | 223 (36.9) | 174 (38.6) | 7 (28.0) | 34 (30.9) | 8 (44.4) | |
| Positive | 373 (61.8) | 272 (60.3) | 18 (72.0) | 73 (66.4) | 10 (55.6) | |
| Donor CMV serostatus (%) | ||||||
| Unknown | 45 (7.5) | 31 (6.9) | 2 (8.0) | 12 (10.9) | 0 (0.0) | |
| Negative | 399 (66.1) | 303 (67.2) | 18 (72.0) | 66 (60.0) | 12 (66.7) | |
| Positive | 160 (26.5) | 117 (25.9) | 5 (20.0) | 32 (29.1) | 6 (33.3) | |
| Graft source, n (%) | ||||||
| BM | 287 (47.5) | 211 (46.8) | 10 (40.0) | 57 (51.8) | 9 (50.0) | |
| PBSC | 309 (51.2) | 235 (52.1) | 14 (56.0) | 51 (46.4) | 9 (50.0) | |
| BM, PBSC | 8 (1.3) | 5 (1.1) | 1 (4.0) | 2 (1.8) | 0 (0.0) | |
| Conditioning intensity, n (%) | ||||||
| Unknown | 23 (3.8) | 19 (4.2) | 1 (4.0) | 3 (2.7) | 0 (0.0) | |
| Nonmyeloablative | 19 (3.3) | 15 (3.5) | 1 (4.2) | 3 (2.8) | 0 (0.0) | |
| Reduced intensity | 70 (12.0) | 52 (12.0) | 3 (12.5) | 12 (11.2) | 3 (16.7) | |
| Myeloablative | 492 (84.5) | 365 (84.3) | 20 (83.3) | 92 (86.0) | 15 (83.3) | |
| T-cell depletion, n (%) | ||||||
| Yes | 198 (33.1) | 155 (34.6) | 7 (29.2) | 30 (27.5) | 6 (33.3) | |
| No | 401 (66.4) | 293 (65.0) | 17 (68.0) | 79 (71.8) | 12 (66.7) | |
| Unknown | 5 (0.8) | 3 (0.6) | 1 (4.0) | 1 (0.9) | 0 (0.0) | |
| HLA match, n (%) | ||||||
| HLA 10/10 matched | 384 (63.6) | 293 (65.0) | 15 (60.0) | 62 (56.4) | 14 (77.8) | |
| HLA 9/10 matched | 220 (36.4) | 158 (35.0) | 10 (40.0) | 48 (43.6) | 4 (22.2) | |
| Year of transplant, n (%) | ||||||
| 2005-2014 | 363 (60.1) | 274 (60.8) | 13 (52.0) | 63 (57.3) | 13 (72.2) | |
| 1989-2005 | 241 (39.9) | 180 (39.2) | 12 (48.0) | 47 (42.7) | 5 (27.8) | |
| Disease status at HCT, n (%) | ||||||
| First complete remission | 247 (40.9) | 179 (39.7) | 10 (40.0) | 49 (44.5) | 9 (50.0) | |
| Second or later complete remission | 156 (25.8) | 118 (26.2) | 5 (20.0) | 25 (22.7) | 8 (44.4) | |
| No remission | 160 (26.5) | 122 (27.1) | 8 (32.0) | 29 (26.4) | 1 (5.6) | |
| Other/unknown | 41 (6.8) | 32 (7.1) | 2 (8.0) | 7 (6.4) | 0 (0.0) | |
| KIR2DS1 HLA-C1/C2 donor, n (%) | ||||||
| KIR2DS1neg/C1/x | 395 (65.4) | 300 (66.5) | 21 (84.0) | 73 (66.4) | 1 (5.6) | |
| KIR2DS1pos/C1/x | 185 (30.6) | 135 (29.9) | 4 (16.0) | 29 (26.4) | 17 (94.4) | |
| C2/C2 | 24 (4.0) | 16 (3.6) | 0 (0.0) | 8 (7.2) | 0 (0.0) | |
BM, bone marrow; CMV, cytomegalovirus; CsA, cyclosporine A; PBSC, peripheral blood stem cell; SD, standard deviation; Tac, tacrolimus; TBI, total body irradiation.
For in vitro NK function studies, unpaired Mann-Whitney U test and 2-way ANOVA with multiple comparisons were used for statistical analysis, and *P ≤ .05 **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001 were used as the P values. Clinical and functional analysis were performed in R and Prism 7 software (GraphPad, San Diego, CA), respectively.
Results
Moderate surface expression of HLA-A*24 and HLA-A*32, but not HLA-A*23 or HLA-A*25, is found on primary lymphocytes
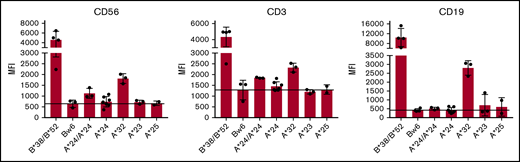
For KIR3DL1+ NK cells from individuals with HLA-B Bw4+ allotypes, titration of NK education and inhibitory strengths is highly correlated with expression levels of both the receptor and ligand.17 We therefore investigated the cell surface expression levels of the different HLA-A Bw4+ allotypes on lymphocytes by staining with anti-HLA-Bw4 antibody PBMCs from healthy donors with HLA-A*24, HLA-A*32, HLA-A*23, or HLA-A*25 genotypes. All donors lacked HLA-B allotypes with the Bw4 epitope, ensuring that any measurable Bw4 staining would be contributed by the HLA-A allotype alone. Of all the HLA-A Bw4+ allotypes, we found HLA-A*32 expression to be most detectable on T, NK, and B cells. Its expression, although detectable, was significantly lower compared with expression of the HLA-B Bw4+ allotypes HLA-B*38 and HLA-B*52, despite the fact that they all belong to the Bw4-I80 subtype group (Figure 1). In addition to HLA-A*32, HLA-A*24 could be detected on the cell surface of T and NK cells, albeit at more modest levels and more so in the context of HLA-A*24 homozygosity. In contrast, cell-surface expression of HLA-A*23 and HLA-A*25 was undetectable on lymphocytes (Figure 1).
HLA-A*24 and HLA-A*32, but not HLA-A*23 or HLA-A*25, are expressed on lymphocytes. PBMCs from healthy donors genotyped for HLA-A*24, HLA-A*32, HLA-A*23, or HLA-A*25 in combination with Bw6/Bw6 or genotyped for HLA-B*38 or HLA-B*52 were stained with fluorescein isothiocyanate-conjugated anti-HLA-Bw4 antibody. Mean fluorescence intensity (MFI) of the indicated Bw4+ HLA-A allotype is depicted among CD3+CD56− (T cells), CD3−CD56+ (NK cells), and CD19+ (B cells) cells.
HLA-A*24 and HLA-A*32, but not HLA-A*23 or HLA-A*25, are expressed on lymphocytes. PBMCs from healthy donors genotyped for HLA-A*24, HLA-A*32, HLA-A*23, or HLA-A*25 in combination with Bw6/Bw6 or genotyped for HLA-B*38 or HLA-B*52 were stained with fluorescein isothiocyanate-conjugated anti-HLA-Bw4 antibody. Mean fluorescence intensity (MFI) of the indicated Bw4+ HLA-A allotype is depicted among CD3+CD56− (T cells), CD3−CD56+ (NK cells), and CD19+ (B cells) cells.
KIR3DL1+ NK cells are educated by HLA-A*24 and HLA-A*32, but not by HLA-A*23 or HLA-A*25
We then investigated how the different HLA-A Bw4+ allotypes contribute to the education of primary KIR3DL1+ NK cells and whether differences in education are evident between the KIR3DL1 high vs low expression subtypes, as occurs with HLA-B Bw4-I80 allotypes.17 Based on previous studies with NK clones suggesting that HLA-A*24 can educate KIR3DL1+ NK cells,22 and on prior binding studies between soluble KIR3DL1 and HLA-A–coated beads,16,17 where HLA-A*32 demonstrated high affinity, HLA-A*24 demonstrated modest affinity, and HLA-A*23 and HLA-A*25 demonstrated either no binding or weak binding, we anticipated that only HLA-A*24 and HLA-A*32 proteins would contribute to NK education. We compared cytotoxic and cytokine response capacities of KIR3DL1high and KIR3DL1low NK cell populations from healthy donors with 1 or more HLA-A alleles encoding the Bw4 epitope. These donors did not have any HLA-B alleles encoding the Bw4 epitope, resulting in exclusive contribution of the Bw4 ligand by HLA-A allotypes only. Cells were evaluated for degranulation and IFN-γ production following culture with the HLA class I-negative K562 cells (supplemental Figure 1 shows the gating strategy).
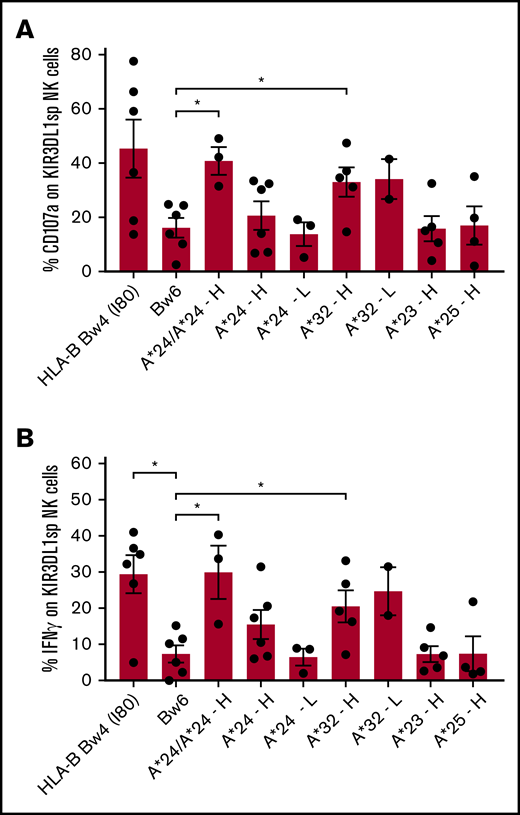
KIR3DL1high NK cells from individuals with HLA-A*32 degranulate and produce more IFN-γ when compared with uneducated KIR3DL1+ NK cells from individuals with no Bw4 epitope (Figure 2A-B), indicating that HLA-A*32 educates KIR3DL1high NK cells for increased responsiveness. KIR3DL1low NK cells from individuals with HLA-A*32 degranulate and produce IFN-γ to similar levels as from individuals with KIR3DL1high NK cells, indicating that KIR3DL1low NK cells are also educated by HLA-A*32. HLA-A*24 also educates KIR3DL1+ NK cells and in a dose-dependent manner; KIR3DL1high NK cells expressing 2 copies of HLA-A*24 showed significantly higher frequency of CD107a mobilization and IFN-γ production compared with uneducated NK cells from individuals lacking Bw4. In comparison, KIR3DL1high NK cells from individuals with only 1 copy of HLA-A*24 displayed intermediate degranulation and IFN-γ production. NK cells from individuals with low expression of KIR3DL1 and 1 copy of HLA-A*24 displayed no appreciable increase in responsiveness over uneducated NK cells (Figure 2A-B). In KIR3DL1high NK cells from individuals with HLA-A*23 and HLA-A*25, no increased degranulation and IFN-γ production was observed over uneducated NK cells (Figure 2A-B), indicating that HLA-A*23 and HLA-A*25 do not educate KIR3DL1+ NK cells. Similar patterns of NK responsiveness were observed when NK cells were cultured with the HLA class I-negative cell line, 721.221 (supplemental Figure 2).
Primary KIR3DL1+NK cells are educated by HLA-A*24 and HLA-A*32, but not HLA-A*23 and HLA-A*25. PBMCs from healthy individuals with HLA-A*24, HLA-A*32, HLA-A*23, or HLA-A*25 and lacking HLA-B Bw4+ alleles were cultured with K562 cells for 5 hours and evaluated for degranulation and cytokine response. CD107a expression (A) and intracellular IFN-γ (B) were evaluated on single positive KIR3DL1 NK cells (KIR3DL1sp). Donor NK cells were stratified according to KIR3DL1high (H) or KIR3DL1low (L) expression. The mean and standard deviation are shown with *P < .05 using unpaired Mann-Whitney U test.
Primary KIR3DL1+NK cells are educated by HLA-A*24 and HLA-A*32, but not HLA-A*23 and HLA-A*25. PBMCs from healthy individuals with HLA-A*24, HLA-A*32, HLA-A*23, or HLA-A*25 and lacking HLA-B Bw4+ alleles were cultured with K562 cells for 5 hours and evaluated for degranulation and cytokine response. CD107a expression (A) and intracellular IFN-γ (B) were evaluated on single positive KIR3DL1 NK cells (KIR3DL1sp). Donor NK cells were stratified according to KIR3DL1high (H) or KIR3DL1low (L) expression. The mean and standard deviation are shown with *P < .05 using unpaired Mann-Whitney U test.
Thus, among the Bw4-bearing HLA-A allotypes, HLA-A*32 and HLA-A*24, but not HLA-A*23 and HLA-A*25, educate primary KIR3DL1+ NK cells for enhanced cytotoxic and cytokine function. For HLA-A*24, however, higher expression of KIR3DL1 and 2 copies of HLA-A*24 are required for detection of increased NK cell responsiveness in vitro. The higher response potential among KIR3DL1+ NK cells from HLA-A*32 individuals is consistent with the higher expression of the HLA allotype, in comparison with the other HLA-A Bw4+ allotypes. Thus, similar to HLA-B Bw4+ allotypes, strength of education is dependent on ligand and receptor densities.17
Primary KIR3DL1+ NK cells are inhibited by HLA-A*24 and HLA-A*32-expressing BLCLs
Inhibition of KIR+ NK cells by HLA ligand in vitro can be predictive of clinical outcomes for patients with solid and hematologic tumors.11,24 Prior in vitro studies examining KIR3DL1+ NK cell inhibition by HLA-A Bw4+ molecules used NK clones, whose function may be altered by long-term culture with cytokine stimulation. We investigated the inhibition potential of HLA-A*24 and HLA-A*32 allotypes by measuring ADCC effector response of KIR3DL1high and KIR3DL1low primary NK cells to BLCLs expressing the HLA-A*24 or HLA-A*32 protein (Table 1; supplemental Figure 3). The degree of inhibition could then be quantitated following the addition of DX9, a KIR3DL1-specific antibody that blocks receptor-ligand engagement. Educated NK cells from 7 Bw4+ healthy donors exhibiting alleles encoding both KIR3DL1high and KIR3DL1low receptor subtypes were analyzed for degranulation in response to Bw4+ BLCLs derived from individuals exhibiting HLA-A*24 or HLA-A*32 and lacking HLA-B Bw4+ alleles by genotype. Any Bw4 epitope detected on the target BLCL was therefore contributed by the HLA-A allele. The anti-CD20 monoclonal antibody rituximab was added to cocultures of NK cells and BLCL target cells as a strong inducer of ADCC, maximizing NK cell activation across the various BLCLs derived from different donors. ADCC-mediated NK activation with the different Bw4-bearing BLCLs was then ascertained by measuring the frequency of CD107a among NK populations single positive for KIR3DL1high or KIR3DL1low receptor expression (Figure 3A) to exclude contribution from other inhibitory NK receptors (supplemental Figure 1). To measure the extent of inhibition signaled by the Bw4 ligand contributed by the different HLA-A–expressing BLCL target cells, cocultures were performed in the presence and absence of the KIR3DL1 blocking antibody DX9 (Figure 3A). Background antibody stimulation of the NK cells could be detected in the absence of BLCL target cells and only in the presence of DX9. We interpret this to be due to binding of rituximab to autologous B cells present in the donor PBMCs, a source of ADCC stimulation that is muted by endogenous Bw4 expression on autologous B cells.
Primary KIR3DL1+NK cells are inhibited by HLA-A*32 and HLA-A*24. PBMCs from 7 healthy Bw4+ donors expressing both KIR3DL1high and KIR3DL1low were incubated for 4 hours with rituximab and BLCL target cells expressing different HLA-A and HLA-B molecules as indicated, in the absence or presence of the anti-KIR3DL1 DX9 antibody. (A) CD107a expression on NK cells singly expressing KIR3DL1high or KIR3DL1low are depicted. Two-way ANOVA with multiple comparisons was used for statistical analysis with **P < .01, ***P < .001, and ****P < .0001. (B) Specific inhibition of KIR3DL1high and KIR3DL1low NK cells by the indicated HLA-A Bw4+ allotypes, expressed as a percent decrease in activation following addition of DX9. Two-way ANOVA with multiple comparisons was used to calculate the significance within the KIR3DL1high (red asterisks) or KIR3DL1low groups (blue asterisks). Comparisons were made between NK cells cultured with Bw6-BLCL (reference) vs either B*38/B*52-BLCL, A*24-BLCL, or A*32-BLCL, respectively, with *P < .05 and ****P < .0001. Comparisons between KIR3DL1high and 3DL1low NK cells within each group are also shown (P = ns [not significant]).
Primary KIR3DL1+NK cells are inhibited by HLA-A*32 and HLA-A*24. PBMCs from 7 healthy Bw4+ donors expressing both KIR3DL1high and KIR3DL1low were incubated for 4 hours with rituximab and BLCL target cells expressing different HLA-A and HLA-B molecules as indicated, in the absence or presence of the anti-KIR3DL1 DX9 antibody. (A) CD107a expression on NK cells singly expressing KIR3DL1high or KIR3DL1low are depicted. Two-way ANOVA with multiple comparisons was used for statistical analysis with **P < .01, ***P < .001, and ****P < .0001. (B) Specific inhibition of KIR3DL1high and KIR3DL1low NK cells by the indicated HLA-A Bw4+ allotypes, expressed as a percent decrease in activation following addition of DX9. Two-way ANOVA with multiple comparisons was used to calculate the significance within the KIR3DL1high (red asterisks) or KIR3DL1low groups (blue asterisks). Comparisons were made between NK cells cultured with Bw6-BLCL (reference) vs either B*38/B*52-BLCL, A*24-BLCL, or A*32-BLCL, respectively, with *P < .05 and ****P < .0001. Comparisons between KIR3DL1high and 3DL1low NK cells within each group are also shown (P = ns [not significant]).
We observed significantly higher frequency of degranulation among KIR3DL1+ NK cells with the DX9 blocking antibody compared with without the blocking antibody when NK cells were cocultured with BLCL target cells expressing HLA-A*24 (P < .001) and HLA-A*32 (P < .0001) (Figure 3A), indicating that, as seen with NK clones, these Bw4-harboring HLA-A allotypes are capable of inhibiting KIR3DL1-expressing primary NK cells (Figure 3A). Interestingly, no difference in degranulation was evident between the KIR3DL1high and KIR3DL1low populations, indicating that both KIR3DL1high and KIR3DL1low NK cells were equally inhibited by HLA-A*24 and HLA-A*32 (Figure 3A-B). Only HLA-A*32 could inhibit NK activation to the same extent as the HLA-B Bw4-I80 allotypes HLA-B*38 and B*52, consistent with the higher expression of the HLA-A allotype and high affinity of the allotype for the receptor.17
Measurement of the inhibitory effect of interaction with HLA-A Bw4+ allotypes on NK activation was calculated as the decrease in NK activation observed in the presence of DX9 compared with NK activation measured in its absence. Among 7 individuals, engagement with HLA-A*32 on the target cell was most inhibitory (Bw6 vs A*32: P < .0001), compared with HLA-A*24, which was mildly inhibitory (Bw6 vs A*24: P < .05) (Figure 3B). Although HLA-A*23 and HLA-A*25 do not contribute to NK education, we tested their ability to inhibit educated KIR3DL1+ cells, finding that HLA-A*23 on BLCL target cells can inhibit modestly and that HLA-A*25 does not inhibit at all (supplemental Figure 4). We conclude that KIR3DL1high and KIR3DL1low NK cells have comparable inhibitory response when engaged by HLA-A*24 and HLA-A*32 and that HLA-A*32 is the strongest KIR3DL1 inhibitor among the HLA-A Bw4+ allotypes with comparable inhibiting strength as HLA-B*38 and HLA-B*52, both Bw4-I80 allotypes.
KIR3DL1/HLA-A*24 is associated with increased post-HCT AML relapse
Strong interacting pairs of HLA-B Bw4+ and KIR3DL1 subtypes are associated with a higher risk of relapse in AML patients who have undergone HLA-matched HCT.11 We reasoned that the educating and inhibitory HLA-A*24 and HLA-A*32 allotypes may also inhibit NK cells in vivo. Because their inhibition of KIR3DL1high and KIR3DL1low cells was comparable, we anticipated that combinations of the HLA-A allotypes with either KIR3DL1 subtype would be inhibitory in vivo, leading to a heightened risk for disease relapse. We performed a retrospective analysis of a cohort of 1729 AML patients who had received a 9/10 or 10/10 HLA allele-matched allograft from an unrelated donor. To assess the impact of Bw4+ epitopes presented by HLA-A allotypes only, patients and donors with HLA-B alleles encoding the Bw4 epitope were excluded. Three patients were removed because of a lack of clinical data, leaving 604 patients for analysis. Among these, 153 donors and patients exhibited the Bw4 epitope contributed by the functionally relevant HLA-A*24 and HLA-A*32, with the remaining 451 patients representing the comparison group. The 1 patient with HLA-A*23 and the 25 patients with HLA-A*25 exclusive of HLA-B Bw4+ alleles were not specifically evaluated because of limited sample size and lack of biological justification, respectively. Patient and donor characteristics are shown in Table 2.
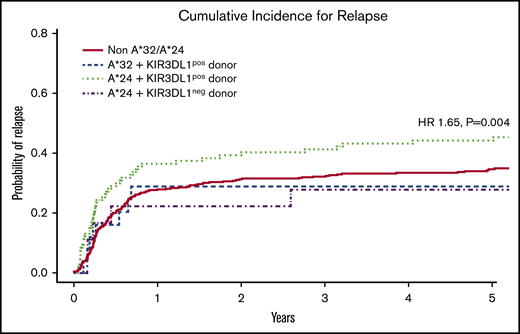
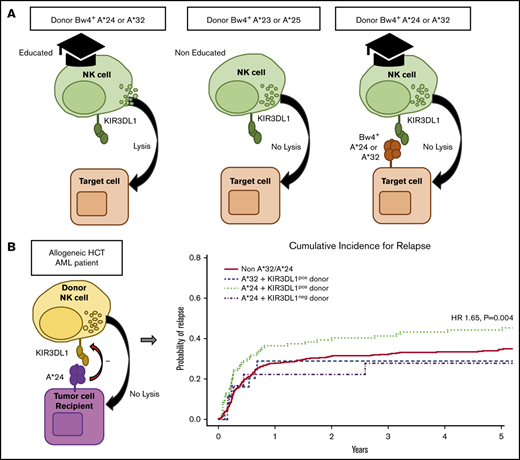
Although the HLA-A*32 molecule exhibits the highest capacity for education and inhibition, we found that the encoding allele occurred within the cohort with low frequency, exclusive of HLA-B Bw4+ alleles (n = 25), limiting informative analysis. In contrast, HLA-A*24 was found to occur at high frequency within the cohort, exclusive of HLA-B Bw4+ alleles (n = 110). Multivariate analysis revealed that the combination of patient HLA-A*24 and donor KIR3DL1 was associated with significantly higher relapse (hazard ratio [HR], 1.65; 95% confidence interval [CI], 1.17-2.32; P = .004) when compared with patients lacking the HLA-A*24 allele or patients exhibiting the HLA-A*24 allele but lacking a KIR3DL1+ donor (Figure 4; Table 3). No difference was found for the risk of mortality (HR, 1.16; 95% CI, 0.91-1.50; P = .235). Nevertheless, this suggests that, similar to the strong interacting HLA-B Bw4+ relationships with KIR3DL1 in HCT, the Bw4-I80 HLA-A*24 allotype on the AML cell can inhibit KIR3DL1+ donor NK cells in vivo, leading to higher risk for relapse.
Cumulative incidence of relapse following HCT in AML patients with inhibitory HLA-A proteins exhibiting the Bw4 epitope. Among patients without contribution of Bw4 from HLA-B alleles, the combination of patient HLA-A*24 with donor KIR3DL1 is associated with higher risk for AML relapse when compared with patients lacking HLA-A*32 and -A*24 (non-A*32/A*24), patients with HLA-A*32 and donor KIR3DL1 (A*32 + KIR3DL1pos donor), and patients with HLA-A*24 but lacking donor KIR3DL1 (A*24 + KIR3DL1neg donor). The KIR3DL1pos donor group includes donors expressing either KIR3DL1high and KIR3DL1low or both. The KIR3DL1neg group consist of donors expressing KIR3DL1null/null, KIR3DL1null/KIR3DS1, and KIR3DS1/3DS1. The indicated HR and P value reflects adjustment for year of transplant, total body irradiation, patient and donor age, conditioning regimen, T-cell depletion, graft type, disease status, cytomegalovirus, sex match, and degree of HLA match. Curve comparisons were completed using Cox proportional hazards regression analysis for the time-to-event post-HCT outcomes.
Cumulative incidence of relapse following HCT in AML patients with inhibitory HLA-A proteins exhibiting the Bw4 epitope. Among patients without contribution of Bw4 from HLA-B alleles, the combination of patient HLA-A*24 with donor KIR3DL1 is associated with higher risk for AML relapse when compared with patients lacking HLA-A*32 and -A*24 (non-A*32/A*24), patients with HLA-A*32 and donor KIR3DL1 (A*32 + KIR3DL1pos donor), and patients with HLA-A*24 but lacking donor KIR3DL1 (A*24 + KIR3DL1neg donor). The KIR3DL1pos donor group includes donors expressing either KIR3DL1high and KIR3DL1low or both. The KIR3DL1neg group consist of donors expressing KIR3DL1null/null, KIR3DL1null/KIR3DS1, and KIR3DS1/3DS1. The indicated HR and P value reflects adjustment for year of transplant, total body irradiation, patient and donor age, conditioning regimen, T-cell depletion, graft type, disease status, cytomegalovirus, sex match, and degree of HLA match. Curve comparisons were completed using Cox proportional hazards regression analysis for the time-to-event post-HCT outcomes.
Multivariate analysis of HLA-A*24 and HLA-A*32 and AML relapse following HCT
| Variable . | N . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| Non-A*32/A*24 | 451 | 1.00 | 1.00-1.00 | NA |
| A*32 + KIR3DL1pos donor | 25 | 0.931 | 0.447-1.942 | .850 |
| A*24 + KIR3DL1pos donor | 110 | 1.649 | 1.172-2.320 | .004 |
| A*24 + KIR3DL1neg donor | 18 | 1.184 | 0.467-3.001 | .722 |
| Variable . | N . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| Non-A*32/A*24 | 451 | 1.00 | 1.00-1.00 | NA |
| A*32 + KIR3DL1pos donor | 25 | 0.931 | 0.447-1.942 | .850 |
| A*24 + KIR3DL1pos donor | 110 | 1.649 | 1.172-2.320 | .004 |
| A*24 + KIR3DL1neg donor | 18 | 1.184 | 0.467-3.001 | .722 |
The number of patients (N), hazard ratios (HR), confidence interval (CI) and P values are shown. KIR3DL1pos donor includes donors expressing either KIR3DL1high, KIR3DL1low, or both. KIR3DL1neg group consist of donors without surface expression of KIR3DL1 (KIR3DL1null/null, KIR3DL1null/KIR3DS1, KIR3DS1/KIR3DS1).
NA, not available.
Discussion
HLA-A Bw4+ allotypes HLA-A*23, HLA-A*24, and HLA-A*32 exhibit different surface expression levels and in vitro capacities to inhibit and educate primary NK cells expressing the cognate KIR3DL1 receptor. We find that primary KIR3DL1+ NK cells from healthy donors are inhibited by HLA-A*24, HLA-A*32, and HLA-A*23, but not by HLA-A*25. These results are in line with previous observations performed using cytokine-expanded NK clones.21-23 Both KIR3DL1high and KIR3DL1low subtypes are inhibited equally by HLA-A*24, despite previous studies demonstrating differential binding of the KIR3DL1 subtypes to HLA-A*24,16,17 confirming that affinity alone is not predictive of inhibitory capacity. Consistent with the in vitro findings of inhibition, the combination of donor KIR3DL1 and patient HLA-A*24 is associated with increased relapse in AML patients following 9/10 and 10/10 HLA-matched allogeneic HCT. This association strongly suggests the in vivo relevance of HLA-A*24 inhibition of KIR3DL1+ NK cells in HCT, where inflammatory conditions lead to HLA class I upregulation on leukemia cells and subsequent NK inhibition.11,28,29 HLA class I antigen expression, including HLA-A, has been described to be preserved on AML blasts,30,31 with the suggestion that HLA-A and HLA-B allotypes expressing the Bw6 epitope is specifically downregulated on leukemic cells,31 leaving HLA allotypes expressing the Bw4 epitope preserved on the surface and potentially heightening the relevance of HLA-A*24 inhibition of KIR3DL1+ NK cells in HCT. No difference for the risk of mortality is found, which in this limited cohort size may be due to increased nonrelapse mortality in the group of interest, however nonsignificant. These results indicate that KIR3DL1/HLA-A*24 should be included with the strong inhibiting KIR3DL1/HLA-B Bw4+ combinations reported previously to be associated with increased risk for relapse.11 For patients with HLA-A*24, selection of donors lacking surface KIR3DL1, such as donors homozygous for KIR3DL1-n or KIR3DS1, may be most appropriate.
HLA-A*24 is found in high frequency within the population across different ethnicities,32 providing adequate power to reveal the significance of the association reported in this study. HLA-A*32, despite encoding a protein with high expression, exhibiting high affinity for KIR3DL1, and mediating high education and inhibition of KIR3DL1+ cells in vitro, could not be confirmed to associate with relapse due to its limited frequency within the patient population. Significantly larger cohorts will be needed to determine if HLA-A*32 in the patient is associated with HCT outcomes.
KIR3DL1-dependent NK clones were derived from an individual expressing HLA-A*24 as the only allele encoding the Bw4+ epitope.22 Using primary NK cells, we can confirm that in addition to HLA-A*24, HLA-A*32 can educate KIR3DL1+ NK cells, but HLA-A*23 and HLA-A*25 cannot. One explanation of why KIR3DL1+ NK cells are inhibited but not educated by HLA-A*23 could be that the ADCC assay is more sensitive to inhibition than activation by K562 cells via NKG2D. Alternatively, in light of our findings that surface density of HLA-A*32 is the highest, and HLA-A*23 and HLA-A*25 the lowest on lymphocytes, we surmise that KIR3DL1+ NK cell education is sensitive to the level of HLA surface density, supporting the model that interactive events contributing to education rely on surface expression of both receptor and ligand. Our finding that KIR3DL1high and KIR3DLlow NK cells are differentially educated by HLA-A*24 provides additional support for this model: KIR3DL1high NK cells are more educated in a homozygous HLA-A*24 environment compared with an environment with only 1 copy of HLA-A*24, and KIR3DL1high cells are more educated than KIR3DL1low cells. The level of HLA-A*24 expression, influenced by the copy number of its allele, may dictate the level of KIR3DL1-mediated education, an interpretation supported by studies demonstrating that copy number of HLA-B Bw4+ and HLA-C2 alleles influences NK education of KIR3DL1+ and KIR2DL1+ NK cells, respectively.17,27,33 Further investigation is needed to confirm this model and will require an accurate understanding of the protein expression of these HLA-A Bw4+ allotypes. Others have reported high allele-specific variability in messenger RNA expression, with HLA-A*24 and HLA-A*23 being the highest expressers and HLA-A*25 and HLA-A*32 the lowest, respectively.34-36 The discrepancy between the published messenger RNA levels and our measurements of surface protein expression is noted; however, the lack of educating capacity of HLA-A*23 and the high educating capacity of HLA-A*32 suggests that protein expression is the more functionally predictive of the 2 measurements.
The same data may contribute to missing-self calculations in HLA-mismatched HCT, such as umbilical or haploidentical HCT. For an ideal NK-mediated graft-versus-leukemia effect, donor NK cells should be educated for reactivity, but infused into a patient lacking the educating KIR ligand.8,9 The finding that HLA-A*24 and HLA-A*32 contribute to education opens the possibility for NK alloreactivity and enhanced leukemia targeting when donor KIR3DL1+ NK cells educated by HLA-A*24 or HLA-A*32 are infused into a patient lacking these and HLA-B Bw4+ ligands.
Graft-versus-host disease, infection, and relapse are the major complications of HCT. Prevention of graft-versus-host disease and treatment of infection have improved because of allograft manipulation, more precise HLA genotyping, and availability of new antimicrobial therapies, leaving disease relapse as the leading cause of death >100 days after allogeneic HCT.37 Our findings demonstrate that HLA-A*24, in combination with KIR3DL1, should be considered in HLA and KIR allele typing for donor selection. Consideration of the KIR3DL1/HLA-A*24 combination may benefit haploidentical HCT outcomes, as well as improve the prognosis among HLA-matched HCT. Adding HLA-A*24 to the previously proposed11 KIR3DL1/HLA-B subtype analysis for refining donor selection algorithms may further reduce relapse and improve survival.
Send data sharing requests via e-mail to the corresponding author, Katharine C. Hsu, at hsuk@mskcc.org.
Acknowledgments
The authors thank Lorna Barnett and Richard O’Reilly (MSKCC) for generating and providing the BLCLs and Mohammed Kazim Panjwani and Rosa Sottile for helpful discussions.
This study was supported by the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (grants U01 AI069197 and R01 AI125651), National Cancer Institute (P01 CA23766), and National Heart, Lung, and Blood Institute (K23 HL140134). Support was also provided by Leukemia & Lymphoma Society and an NIH, National Cancer Institute core grant (P30 CA008748) to Memorial Sloan Kettering Cancer Center.
Authorship
Contribution: K.v.d.P., J.-B.L.L., and K.C.H. created and designed the study; B.C.S. analyzed the patient cohort; T.A.G. and P.A.S. provided statistical analysis; K.v.d.P., J.-B.L.L., B.C.S., S.P., P.A.S., T.A.G., E.W.P., and K.C.H. collected and assembled the data; K.v.d.P., J.-B.L.L., B.C.S., P.A.S., and K.C.H. analyzed and interpreted the data; K.v.d.P. drafted the manuscript; and all authors reviewed, revised, and approved the final manuscript.
Conflict-of-interest disclosure: K.C.H. has a patent application on the KIR3DL1 multiplex polymerase chain reaction assay used in this study and patent applications pertaining to the selection of hematopoietic cell donors on the basis of KIR and HLA for hematopoietic cell transplantation; she also has had consulting roles for Rubius Therapeutics. T.A.G. holds stock in Johnson & Johnson and has a consulting role for Nohla Therapeutics. The remaining authors declare no competing financial interests.
The current affiliation for K.v.d.P. is Department of Medicine, Stanford University, Stanford, CA.
Correspondence: Katharine C. Hsu, Department of Medicine, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: hsuk@mskcc.org.
References
Author notes
The full-text version of this article contains a data supplement.

![Primary KIR3DL1+NK cells are inhibited by HLA-A*32 and HLA-A*24. PBMCs from 7 healthy Bw4+ donors expressing both KIR3DL1high and KIR3DL1low were incubated for 4 hours with rituximab and BLCL target cells expressing different HLA-A and HLA-B molecules as indicated, in the absence or presence of the anti-KIR3DL1 DX9 antibody. (A) CD107a expression on NK cells singly expressing KIR3DL1high or KIR3DL1low are depicted. Two-way ANOVA with multiple comparisons was used for statistical analysis with **P < .01, ***P < .001, and ****P < .0001. (B) Specific inhibition of KIR3DL1high and KIR3DL1low NK cells by the indicated HLA-A Bw4+ allotypes, expressed as a percent decrease in activation following addition of DX9. Two-way ANOVA with multiple comparisons was used to calculate the significance within the KIR3DL1high (red asterisks) or KIR3DL1low groups (blue asterisks). Comparisons were made between NK cells cultured with Bw6-BLCL (reference) vs either B*38/B*52-BLCL, A*24-BLCL, or A*32-BLCL, respectively, with *P < .05 and ****P < .0001. Comparisons between KIR3DL1high and 3DL1low NK cells within each group are also shown (P = ns [not significant]).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/4/19/10.1182_bloodadvances.2020002086/2/m_advancesadv2020002086f3.png?Expires=1767906139&Signature=bFAJlwM068zL09GlcQc0OL7KUGJuBjOEu6SusXSN49-ZZWpD52q-lFg6BYfRqCoF6lKxmGVtXXMCpiPxobTKQY7UrJ4mJnQrRAD7D8yV8EPOGQq1Wf892B7wKAMq6okdDCPcLWI13ZoW2Qb1U8O3u6hb7cUWlnTrZMLqKTtIFGG7qscY7a20nFuI32V1C927w3dYuyircrZ77UHrjfdG8RSPZde5-7bFMkkXjoDWscfbHSGd8ALHw4EvEcpBYuYzrviQsGISxskNmsqq1MMl~KBnYD7p982Hq~rUA04oT-yPF4TNRwcuYH~sZbz~Ux80eQoWOMzG-AAg5K0r9~Zxpg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)