Key Points
The integrin Mac-1 I-domain interaction with platelet receptor GPIbα has been mapped by NMR.
The GPIbα LRR capping α-helix coordinates directly to the Mac-1 MIDAS Mg2+ ion via an acidic residue.
Abstract
Cell-surface receptor interactions between leukocyte integrin macrophage-1 antigen (Mac-1, also known as CR3, αMβ2, CD11b/CD18) and platelet glycoprotein Ibα (GPIbα) are critical to vascular inflammation. To define the key residues at the binding interface, we used nuclear magnetic resonance (NMR) to assign the spectra of the mouse Mac-1 I-domain and mapped the residues contacting the mouse GPIbα N-terminal domain (GPIbαN) to the locality of the integrin metal ion-dependant adhesion site (MIDAS) surface. We next determined the crystal structures of the mouse GPIbαN and Mac-1 I-domain to 2 Å and 2.5 Å resolution, respectively. The mouse Mac-1 I-domain crystal structure reveals an active conformation that is stabilized by a crystal contact from the α7-helix with a glutamate side chain completing the octahedral coordination sphere of the MIDAS Mg2+ ion. The amino acid sequence of the α7-helix and disposition of the glutamic acid matches the C-terminal capping region α-helix of GPIbα effectively acting as a ligand mimetic. Using these crystal structures in combination with NMR measurements and docking analysis, we developed a model whereby an acidic residue from the GPIbα leucine-rich repeat (LRR) capping α-helix coordinates directly to the Mac-1 MIDAS Mg2+ ion. The Mac-1:GPIbαN complex involves additional interactions consolidated by an elongated pocket flanking the GPIbαN LRR capping α-helix. The GPIbαN α-helix has an HxxxE motif, which is equivalent by homology to RxxxD from the human GPIbαN. Subsequent mutagenesis of residues at this interface, coupled with surface plasmon resonance studies, confirmed the importance of GPIbαN residues H218, E222, and the Mac-1 MIDAS residue T209 to formation of the complex.
Introduction
The integrin receptor macrophage-1 antigen (Mac-1, also known as CR3, integrin αMβ2, or CD11b/CD18) is expressed on the surface of myeloid leukocytes and mediates numerous responses of these cells critical to innate immunity.1 The Mac-1 receptor contributes to the recruitment, firm adhesion, and transendothelial migration of leukocytes at sites of vascular injury and facilitates tissue inflammation.2 Biochemical and cell-based studies have characterized the interactions of the Mac-1 integrin with diverse ligands, including plasma protein fibrinogen,3 complement protein fragment iC3b,4 high-molecular-weight kininogen ,5 and the cell-surface receptors platelet glycoprotein Ibα (GPIbα)2 and intercellular adhesion molecule 1 (ICAM-1).6 The Mac-1 integrin exists in an inactive conformation and, when activated by a variety of stimuli, undergoes a structural change to an active form capable of binding to ligands with high affinity.7,8 Concurrently, allosteric changes occur on ligand binding that result in “outside-in” cell signaling.9
Mac-1–deficient mice show delayed thrombosis, but largely unimpaired hemostasis.10 Blocking the Mac-1:GP1bα interaction prevents neutrophil extracellular trap formation,11 and an antibody targeting Mac-1 was shown to block inflammation.12 Inhibiting the Mac-1 I-domain with the small molecule allosteric regulator leukaderin-1 has demonstrated efficacy in animal models of inflammatory disease.13,14
For the majority of Mac-1 interactions described, the inserted αM-subunit, or “I-domain,” is the principal ligand-binding domain. The Mac-1 I-domain contains a Mg2+ binding site on its surface at the top of the β sheet called the metal ion-dependant adhesion site (MIDAS), which is capable of accepting the side chain of an acidic residue (aspartate or glutamate) from coordinating ligands.1,7 A crystal structure has defined the interaction between Mac-1 and iC3b,4 but the complex between Mac-1 and GPIbα and the principal ligand fibrinogen3 are poorly understood at the molecular level. Here, nuclear magnetic resonance (NMR) assignment of the mouse Mac-1 I-domain, together with X-ray structural analysis of both the mouse GP1bαΝ and Mac-1 I-domain, has enabled us to map the binding surface and model the interaction site for the GP1bα leucine-rich repeat (LRR) N-terminal domain (GP1bαN) with the MIDAS face of Mac-1.
Materials and methods
Protein expression, purification, and characterization
A complementary DNA fragment encoding mouse GP1bαN residue E1 to T266 (mature protein sequence numbering) was cloned into the pMT-puro vector for expression using the DES system (Invitrogen). At the N terminus, the signal sequence corresponds to a Drosophila homolog of the immunoglobin binding chaperone protein secretion signal, and a polyhistidine tag sequence TRTGHHHHHH was added at the C terminus. Site-specific mutations H218A and E222A were introduced using the QuikChange method (Stratagene) and confirmed by DNA sequencing. The mouse amino acid sequence numbering is defined by alignment with the mature human GP1bα sequence, such that H218 corresponds to H234 in the UniProt numbering of entry GP1BA_MOUSE, which includes the signal sequence. Drosophila S2 cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum at 28°C and transfection was performed in each case using calcium phosphate. Cells were grown for an additional 48 hours before selection with puromycin to establish stable cell lines. Serum-free Express Five (Invitrogen) insect culture media was collected containing secreted proteins. Initial capture of the crude product was performed using Ni-sepharose affinity chromatography, followed by gel filtration and ion exchange chromatography (supplemental Figure 1). Protein identity and successful removal of the signal peptide was confirmed by trypsin digest mass spectrometry, analyzed using the Mascot Database (ProteinID, University of York) shown in supplemental Figures 1 and 2.
The mouse Mac-1 I-domain gene fragment (P129 to A318; mature protein sequence numbering) was cloned into the pGEX-4T-1 vector, for expression as an N-terminal GST-fusion using Escherichia coli. All mutants were produced using Q5 site-directed mutagenesis. XL1-Blue and BL21 cells were transformed using calcium chloride for storage and expression, respectively. BL21 cells were grown in LB media (for surface plasmon resonance [SPR] and mass spectrometry) or M9 minimal media (for NMR spectroscopy) supplemented with 15N ammonium chloride and/or 13C glucose as required for isotope labeling. GST-affinity chromatography and incubation with thrombin resulted in the separation of the tag (residual Gly-Ser remains at the N terminus) and further purification using cationic exchange chromatography. Mass spectrometry, gel filtration studies, NMR line width analysis, and diffusion measurements indicate that the recombinant Mac-1 I-domain is monomeric in solution (supplemental Figures 3 and 4).
NMR spectroscopy
NMR data were recorded at 25°C on a Bruker 800 MHz Avance III spectrometer with a QCI cryoprobe using Topspin 3.1 software. Backbone resonances (13C/15N) were assigned using HNCO, HN(Cα)CO, CβCαNH, and CβCα(CO)NH experiments. Backbone assignment (>146/182 residues) was facilitated by a number of locally perturbing single-point mutations (to Ala/Gly) and through selective unlabeling experiments (Glu and Lys, in particular). 15N-TROSY 2D spectra were collected in titration experiments at a Mac-1 concentration of 120 μM in 25 mM potassium phosphate buffer (pH 7.0), 25 mM NaCl, 5% D2O, and 0.02% NaN3 using 1.5 equivalents of MgCl2 to ensure metal complexation. 15N-TROSY spectra were collected at 0.2 molar ratio intervals up to a 2:1 excess of GPIbαN. Chemical shift perturbations (CSPs) were calculated using a weighting of 1H and 15N shifts according to: CSP = √ [1/2 × (δH2 + 0.14 × δN2)]. For structural modeling, the standard deviation (SD) from the mean CSP was calculated and mean + SD and mean + 2 × SD used for surface mapping.
Quantitation of Mac-1 I-domain ligand binding
SPR was performed using a BIAcore 3000 (GE Healthcare). Ligands of recombinant mouse GPIbαN, GPIbαN variants H218A and E222A, and fibrinogen (σ) were amine-coupled to a CM5 sensor chip (GE Healthcare); a reference cell was prepared by blank amine-coupling. Recombinant Mac-1 I-domain wild-type and T209A variant samples at protein concentrations ranging from 3.125 μM to 200 μM, in running buffer (10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 140 mM NaCl, 1 mM MgCl2) were injected at a flow rate of 50 μL/min. Kinetic studies of the Mac-1 I-domain wild-type and T209A constructs were conducted as a dilution series, in the presence and absence of Mg2+ against the immobilized GPIbαN. Each of the series contains a minimum of 10 concentrations (11.25-200 µM) and was repeated a minimum of 2 times. The Mg2+ ion dependence of the GPIbαN interaction was tested against a wild-type Mac-1 I-domain concentration series (6-100 µM) in which the running buffer contained 5 mM EDTA to sequester metal ions. Binding to the immobilized GPIbαN was also evaluated using a dilution series of the antibody Xia.B2 (emfret ANALYTICS) with protein concentration of 3 to 50 nM. The CM5 chip was regenerated with 2 M NaCl after each experiment. Binding curves were generated by subtracting the appropriate controls and the resulting curves were analyzed using BIAevaluation software or a steady-state equilibrium approach using a Hill plot generated with Prism 6 (GraphPad Software Inc.).
Protein crystallization and crystal structure determination
Recombinant mouse GP1bαN was concentrated to 12 mg/mL and subjected to sparse matrix screens of Morpheus, Proplex, Index, PACT, JCSG+, and MIDAS using the sitting drop vapor diffusion method. Crystals of GP1bαN grew in Morpheus condition D2 containing 0.2 M mixture of 1,6-hexanediol; 1-butanol; 1,2-propanediol; 2-propanol; 1,4-butanediol; 1,3-propanediol, 0.1 M imidazole; MES monohydrate (acid) pH 6.5, 20% v/v ethylene glycol; and 10% w/v PEG 8000. Crystals of the mouse Mac-1 I-domain (6 mg/mL) grew in the JCSG+ screen condition D7, 0.2 M lithium sulfate, 0.1 M tris(hydroxymethyl)aminomethane, pH 8.5, 40% PEG 400. Crystals were cryocooled in liquid nitrogen and X-ray diffraction data were collected using the ID30B beamline at the ESRF (GP1bαN) and Diamond beamline I24 (Mac-1 I-domain). Data were processed with XDS and molecular replacement was performed with PHASER using a human GP1bαN15 and Mac-1 I-domain structure7 (Table 1). Partial model building was performed using BUCCANEER and completed manually using COOT and refined with REFMAC5 (deposited PDB code: 6EJX).
Molecular docking calculations
Three-dimensional models of the Mac-1 I:GPIbαN docked complex structures were calculated using HADDOCK16 with template Mac-1 I-domain (PDB code: 1IDO) and GPIbαN (PDB code: 1M0Z) crystal structures used. Active residues were defined based on the Mac-1 I-domain NMR titration experiments: E244, G143, and N147 plus the MIDAS Mg2+ ion. Active residues for GPIbαN were D222 and R218.
Results
NMR assignment and structural analysis of the Mac-1 I-domain
A bacterially expressed mouse Mac-1 I-domain construct (residues 129-318) was isotopically (13C/15N) labeled and the NMR spectra assigned using heteronuclear 3-dimensional NMR experiments. The 1H/15N heteronuclear single quantum coherence is highly dispersed with sharp resonances indicative of well-ordered secondary structure in both the Mg2+ free and bound state (supplemental Figure 5). We also prepared and characterized 2 Mac-1 mutants, T209A and F302W, which have previously been reported as “inactive” (T209A) and “pseudo-open” (F302W) variants of the I-domain.17,18 The side chain of T209 coordinates directly with the Mg2+ ion only in the “open” conformation of the Mac-1 I-domain,4,7 and previous crystallographic studies have defined a large allosteric conformational shift between “open” and “closed” states, which links the switching of the coordination of the metal ion from D140 to T209 to a shift in the position of helix α7.7,19 T209 has also been shown to play a direct and critical role in Mac-1 ligand binding to ICAM-1 and iC3b.6 The CD spectra of the mutants and wild-type Mac-1 are superimposable, showing no significant changes in protein secondary structure and folding (supplemental Figure 1B). NMR of the 15N-labeled mutant proteins confirms retention of the core structure; however, extensive perturbations across the MIDAS face are evident in both mutants (supplemental Figure 5).
Crystallographic data collection and refinement statistics
| Sample . | Mouse GP1bαN . | Mouse Mac-1 I . |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P41212 |
| Cell dimensions | ||
| a, b, c (Å) | 61.5, 72.8, 164.0 | 62.9, 62.9, 336.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution, Å | 29.9-2.0 | 45.9-2.5 |
| Rmerge* | 11.1 | 12.5 |
| I/σI; CC(1/2)† | 7.0 (1.9); 0.997 (0.78) | 8.2 (1.5); 0.903 (0.68) |
| Completeness (%)† | 99.0 (90.0) | 86.7 (67.2) |
| Redundancy† | 4.3 (1.3) | 6.2 (2.5) |
| Refinement | ||
| Number of reflections | 48095 | 21236 |
| Rwork/Rfree‡ | 0.192/0.237 | 0.222/0.271 |
| B factors, Å2 | ||
| Protein | 29.8 | 48.2 |
| RMS deviations | ||
| Bond lengths, Å | 0.018 | 0.021 |
| Bond angles, ° | 1.96 | 1.85 |
| Sample . | Mouse GP1bαN . | Mouse Mac-1 I . |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P41212 |
| Cell dimensions | ||
| a, b, c (Å) | 61.5, 72.8, 164.0 | 62.9, 62.9, 336.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution, Å | 29.9-2.0 | 45.9-2.5 |
| Rmerge* | 11.1 | 12.5 |
| I/σI; CC(1/2)† | 7.0 (1.9); 0.997 (0.78) | 8.2 (1.5); 0.903 (0.68) |
| Completeness (%)† | 99.0 (90.0) | 86.7 (67.2) |
| Redundancy† | 4.3 (1.3) | 6.2 (2.5) |
| Refinement | ||
| Number of reflections | 48095 | 21236 |
| Rwork/Rfree‡ | 0.192/0.237 | 0.222/0.271 |
| B factors, Å2 | ||
| Protein | 29.8 | 48.2 |
| RMS deviations | ||
| Bond lengths, Å | 0.018 | 0.021 |
| Bond angles, ° | 1.96 | 1.85 |
RMS, root mean square.
Rmerge = Sum(h) [Sum(j) [I(hj) − <Ih>]/Sum(hj) <Ih>, where I is the observed intensity and <Ih> is the average intensity of multiple observations from symmetry-related reflections calculated with SCALA.
Values in parentheses are for the highest resolution shell.
Rwork = Sum(h) ||Fo|h − |Fc|h| / Sum(h)|Fo|h, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree computed as in Rwork, but only for 5% randomly selected reflections, which were omitted in refinement, calculated using REFMAC.
NMR analysis of the Mac-1 I-domain interaction with GP1bαN
Recombinant mouse GP1bαN was expressed and purified from insect cells (supplemental Figures 1 and 2). NMR chemical shifts (1H/15N) are particularly sensitive to changes in chemical environment and the conformation associated with ligand binding and were used to characterize the GP1bαN interaction with the Mac-1 I-domain. A buffered solution of GP1bαN was titrated into a 120 μM solution of 15N-Mac-1 I-domain in the presence of 1.5 molar equivalents of Mg2+, and 1H/15N heteronuclear single quantum coherence spectra were collected over a range of ligand concentrations up to a Mac-1 I:GP1bαN molar ratio of 1:2.
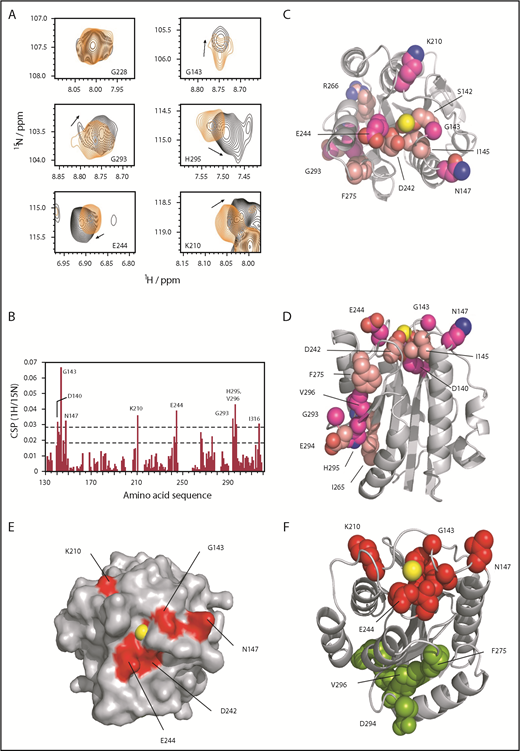
A significant number of fast-exchange CSPs are observed in response to ligand binding. However, a number of residues within the Mac-1 I-domain C-terminal region (helix α7) showed some evidence of signal broadening and peak distortion in the bound state. Statistically significant CSP effects are identified for 16 residues of the Mac-1 I-domain (Figure 1) and enable us to identifying 2 well-defined binding patches. The first group of residues, which are in close proximity or intimately involved in MIDAS Mg2+ coordination, includes D140, S142, G143, I145, and D242 together with the adjacent solvent-exposed residues N147, K210, and E244. Residues D242, E244, and D273 cluster on the surface of the I-domain to form a negatively charged patch that is adjacent to the MIDAS. The second group of Mac-1 residues I265, R266, F275, G293, E294, H295, and V296 (Figure 1D-E) are distal to the MIDAS and appear to be linked to it through conformational changes propagated away from the metal center.
Mapping the Mac-1 I-domain binding site with GP1bαN by NMR. (A) Selected peaks showing CSP effects between bound and unbound (the first panel for G228 is shown as a control, in which no perturbation is observed). (B) Representation of CSP effects, with dashed lines showing statistically significant perturbation (lower line, mean CSP +1 SD; upper line, mean CSP +2 SD) residues above the latter. (C) CSP effects mapped to the Mac-1 surface with residues in darker pink corresponding to mean +2 SD, top-down view onto the MIDAS (Mg2+ shown in yellow). (D) Side view showing distal CSP effects removed from the MIDAS site (top). (E) Surface representation showing the binding patch around the MIDAS with key residues in red. (F) Differentiation between the MIDAS binding patch, shown in red, and those distal residues (in green) affected through allosteric changes from binding GP1bαN at the MIDAS site (a few key residues are highlighted).
Mapping the Mac-1 I-domain binding site with GP1bαN by NMR. (A) Selected peaks showing CSP effects between bound and unbound (the first panel for G228 is shown as a control, in which no perturbation is observed). (B) Representation of CSP effects, with dashed lines showing statistically significant perturbation (lower line, mean CSP +1 SD; upper line, mean CSP +2 SD) residues above the latter. (C) CSP effects mapped to the Mac-1 surface with residues in darker pink corresponding to mean +2 SD, top-down view onto the MIDAS (Mg2+ shown in yellow). (D) Side view showing distal CSP effects removed from the MIDAS site (top). (E) Surface representation showing the binding patch around the MIDAS with key residues in red. (F) Differentiation between the MIDAS binding patch, shown in red, and those distal residues (in green) affected through allosteric changes from binding GP1bαN at the MIDAS site (a few key residues are highlighted).
Crystal structures of the mouse GP1bαN and Mac-1 I-domain
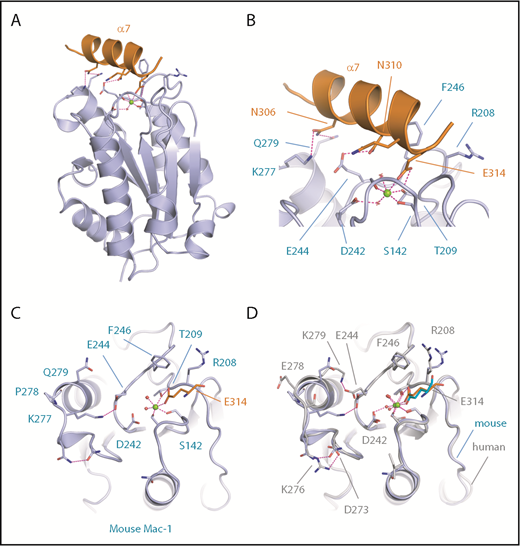
We next determined the crystal structure of mouse Mac-1 I-domain to 2.5 Å resolution. This revealed the mouse Mac-1 I-domain in an open conformation with the MIDAS metal ion coordinated by T209, S142, S144, and a glutamic acid residue (E314) ligand mimetic supplied by a crystal contact from the C-terminal helix (α7) of the I-domain (orange in Figure 2A; supplemental Video 1). The C-terminal Mac-1 I-domain α-helix consists of the amino acid sequence 310-NQLQE-314, which resembles the sequence from the α-helix in the GPIbαN capping region spanning residues 218-HWLQE-222, and is equivalent structurally in terms of the α-helical disposition of the glutamic acid residue. Figure 2B shows that the α-helix sits in a groove on the surface of the Mac-1 I-domain packing against residues F246 and R208, forming multiple hydrogen bonded contacts between the sidechains of N310 and N306 and the sidechains of E244 and K277 on the MIDAS face. For comparison, crystal structures are available for the human Mac-1 I-domain as an open conformation in complex with ligand iC3b,4 simvastatin,20 and stabilized by a crystal contact.7,8 The mouse and the human Mac-1 I-domain crystal structures are superposed in Figure 2D, revealing the MIDAS metal ion coordinating acidic residue consistently occupies an identical position simultaneously bonding to both the metal ion and the hydroxyl of T209. Subtle differences occur across the MIDAS face with the introduction of K277 in the mouse Mac-1 forming a salt bridge to E244, replacing the K279-E244 salt bridge observed in the human structure (Figure 2C).
Crystal structure of the mouse Mac-1 I-domain. (A) A cartoon diagram is shown for the crystal structure of the mouse Mac-1 I-domain (purple) with the crystal contact stabilizing the active conformation derived from the C-terminal helix α7 in orange. The MIDAS bound Mg2+ ion is shown as a sphere (green) and key residues are shown as sticks and electrostatic interactions as dashed magenta lines. (B) A close-up view of the MIDAS site with residues from helix α7 labeled. (C) A second view of the Mac-1 I-domain MIDAS site with the crystal contact residue E314 shown coordinating the Mg2+ ion and the sidechain hydroxyl of T209. (D) Superposition of the human (gray) and mouse (purple) crystal structures in the region of the MIDAS face. The stabilization glutamic acid is shown as sticks in cyan (human) and orange (mouse).
Crystal structure of the mouse Mac-1 I-domain. (A) A cartoon diagram is shown for the crystal structure of the mouse Mac-1 I-domain (purple) with the crystal contact stabilizing the active conformation derived from the C-terminal helix α7 in orange. The MIDAS bound Mg2+ ion is shown as a sphere (green) and key residues are shown as sticks and electrostatic interactions as dashed magenta lines. (B) A close-up view of the MIDAS site with residues from helix α7 labeled. (C) A second view of the Mac-1 I-domain MIDAS site with the crystal contact residue E314 shown coordinating the Mg2+ ion and the sidechain hydroxyl of T209. (D) Superposition of the human (gray) and mouse (purple) crystal structures in the region of the MIDAS face. The stabilization glutamic acid is shown as sticks in cyan (human) and orange (mouse).
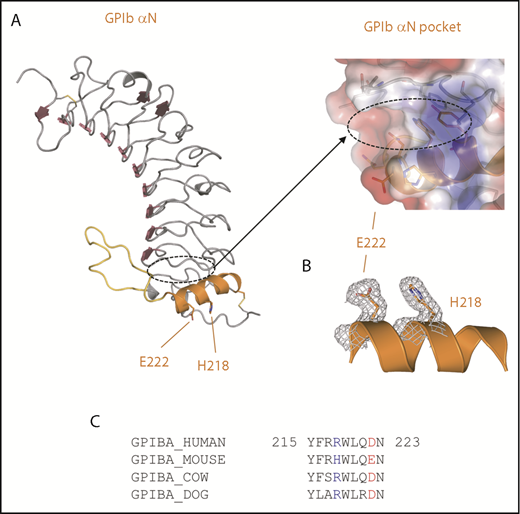
We then determined the crystal structure of the mouse GP1bαN to 2.0 Å resolution to examine the conformation of the LRR C-terminal capping α-helix, which is important for the interaction with Mac-1.2 This structure reveals overall features of the LRR solenoid flanked by N- and C-terminal disulphide-bonded capping regions and R-loop, as observed previously for the human GP1bαN structure.21 The junction between the C-terminal capping region α-helix and the LRR solenoid forms an elongated pocket (Figure 3A, dotted elliptical line). In a notable departure from the human sequence, Figure 3B-D reveals that the mouse GP1bαN has the substitutions H218 and E222 that, by analogy with the human GP1bαN structure, are also surface exposed (Figure 3B). A comparison of structures shows that mouse GP1bαN H218 and E222 occur with the same disposition as human GP1bαN R218 and D222, respectively, and a sequence alignment of this region reveals that charged residues at these positions are conserved across other mammalian species (Figure 3C).
Crystal structure of the mouse GP1bαN. (A) A cartoon diagram is shown for the crystal structure of the mouse GPIbαN with the C-terminal LRR capping α-helix colored orange and R-loop colored yellow. GPIbα residues H218 and E222 involved in the interaction with Mac-1 are shown as sticks. The elongated pocket formed in the GPIbα LRR capping region is indicated by a dashed line and represented as a transparent charged surface with key residues lining the pocket shown as sticks. (B) Electron density (2Fo-Fc) from the crystal structure of the mouse GPIbαN C-terminal capping α-helix residues H218 and E222 (gray mesh). (C) Amino acid sequence alignment of GPIbα residues 215 through 223 (human sequence numbering without the signal sequence) from the GPIbα LRR capping α-helix for human, mouse, dog, and cow. Key charged residues 218 and 222 are colored blue and red, respectively.
Crystal structure of the mouse GP1bαN. (A) A cartoon diagram is shown for the crystal structure of the mouse GPIbαN with the C-terminal LRR capping α-helix colored orange and R-loop colored yellow. GPIbα residues H218 and E222 involved in the interaction with Mac-1 are shown as sticks. The elongated pocket formed in the GPIbα LRR capping region is indicated by a dashed line and represented as a transparent charged surface with key residues lining the pocket shown as sticks. (B) Electron density (2Fo-Fc) from the crystal structure of the mouse GPIbαN C-terminal capping α-helix residues H218 and E222 (gray mesh). (C) Amino acid sequence alignment of GPIbα residues 215 through 223 (human sequence numbering without the signal sequence) from the GPIbα LRR capping α-helix for human, mouse, dog, and cow. Key charged residues 218 and 222 are colored blue and red, respectively.
Docking of the GPIbαN and Mac-1 I-domain 3-dimensional structures
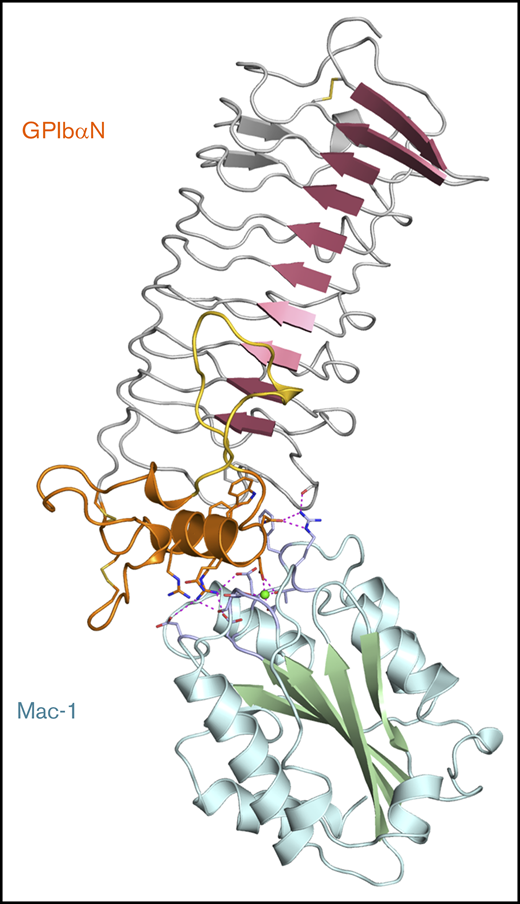
Previous cell-based and biochemical studies identified residues 218 through 228 from GP1bα as the principal determinants of the Mac-1 interaction2 encompassing the human GP1bαN LRR C-terminal capping α-helix, but did not specify whether a critical acidic residue was involved. Studies using a variety of techniques implicated Mac-1 as residing on the MIDAS face and residues T213 and R216 as being located away from the MIDAS face, but did not propose a role for the Mg2+ ion or an overall model for the complex with GP1bαN.10,22 We thus set out to investigate this by generating models of the Mac-1 I:GP1bαN complex using the program HADDOCK.16 Multiple crystal structures are available for the human Mac-1 I-domain in the open conformation4,7,8,18,23 and human GP1bαN domain,15,21,24-26 and they were used as templates for docking.
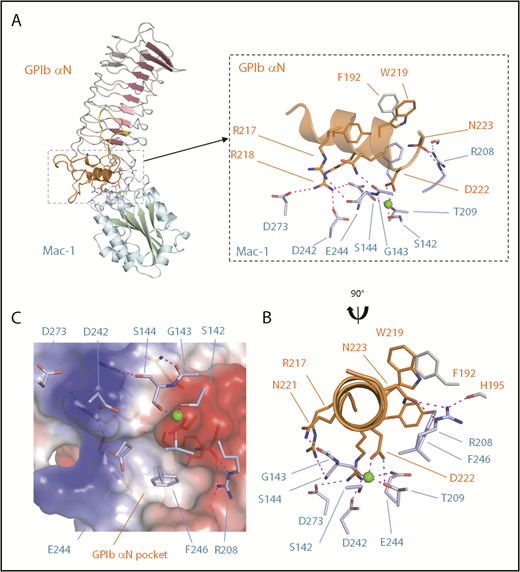
Docking was performed using NMR distance restraints between the Mac-1 MIDAS Mg2+ ion and residues G143, N147, D242, and E244, which were identified as having significant CSP effects in the NMR titration experiments, together with GP1bαN surface-exposed residues R218, D222, and N223.2 The top scoring structure and details of the interface are shown in Figure 4A. The Mac-1 I:GP1bαN complex buries a surface area of 593 Å2 (calculated using PISA), and 2 key features that define the unique orientation are that GP1bαN D222 coordinates to the Mac-1 MIDAS Mg2+ ion and GP1bαN R218 forms bidentate salt bridges with Mac-1 residues D242 and E244 (supplemental Videos 2 and 3). As viewed down the central projection of the GP1bαN capping α-helix, colored orange in Figure 4B, residues G143 and S144 of Mac-1 (lower left) hydrogen bond to the side chains of N221 and R217 of GP1bα, respectively. Flanking interactions also occur on the right of Figure 4B as R208 from Mac-1 forms hydrogen bonds to the side chains of N223 of GP1bα, and its own H195 main chain carbonyl. The Mac-1 I-domain F246 side chain inserts into an elongated hydrophobic pocket in the GP1bαN LRR capping region defined by GP1bα residues F192, S194, Y215, and W219 (Figure 4C).
The Mac-1 I:GP1bαN complex. (A) A cartoon diagram of the docked complex of the crystal structures of human GPIbαN and the Mac-1 I-domain with the Mac-1 secondary structures colored blue/green and GPIbαN colored red/orange. Boxed is a close-up view of the interface where the Mg2+ ion bound to the Mac-1 MIDAS site is shown as a sphere and electrostatic interactions are shown as dashed purple lines. The GPIbαN C-terminal LRR capping region is colored orange and residue F192 from the LRRs is in gray. (B) A second view related by a 90° rotation. GPIbαN residue D222 coordinates the Mac-1 MIDAS Mg2+ ion and surrounding residues on the MIDAS surface (light blue). (C) Charged surface representation of the GPIbαN LRR capping region showing an elongated pocket formed by GPIbα residues Y215, W219, and F192 is flanked by a region of positive charge (GPIbα R218) and negative charge (GPIbα D222) that form complementary interactions with Mac-1 residues shown as sticks (light blue).
The Mac-1 I:GP1bαN complex. (A) A cartoon diagram of the docked complex of the crystal structures of human GPIbαN and the Mac-1 I-domain with the Mac-1 secondary structures colored blue/green and GPIbαN colored red/orange. Boxed is a close-up view of the interface where the Mg2+ ion bound to the Mac-1 MIDAS site is shown as a sphere and electrostatic interactions are shown as dashed purple lines. The GPIbαN C-terminal LRR capping region is colored orange and residue F192 from the LRRs is in gray. (B) A second view related by a 90° rotation. GPIbαN residue D222 coordinates the Mac-1 MIDAS Mg2+ ion and surrounding residues on the MIDAS surface (light blue). (C) Charged surface representation of the GPIbαN LRR capping region showing an elongated pocket formed by GPIbα residues Y215, W219, and F192 is flanked by a region of positive charge (GPIbα R218) and negative charge (GPIbα D222) that form complementary interactions with Mac-1 residues shown as sticks (light blue).
SPR analysis of the Mac-1 I:GPIbαN interaction
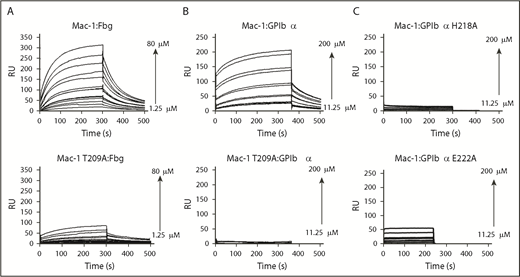
To further evaluate the contribution of amino acid side chains at the center of the proposed mouse Mac-1 I:GP1bαN interface, we used site-directed mutagenesis coupled with SPR. To establish the assay, the SPR response was first observed with Mac-1 I-domain analyte and immobilized full-length fibrinogen, which has been characterized in detail from previous studies.3 SPR analysis (Figure 5A) shows a high-affinity interaction with evidence of multiple binding modes; an estimated Kd of 30 μM was derived from the equilibrium values of response units was consistent with previous reports.3
SPR analysis of Mac-1 I-domain interactions. Plots of SPR sensorgrams measured in RU on the y-axis illustrating wild-type Mac-1 (top) or Mac-1 T209A (bottom) binding to fibrinogen (Fbg; A) and GP1bαN (B), and shown in wild-type Mac-1 I-domain binding to the GP1bαN mutants H218A (C, top) and E222A (C, bottom). RU, response unit.
SPR analysis of Mac-1 I-domain interactions. Plots of SPR sensorgrams measured in RU on the y-axis illustrating wild-type Mac-1 (top) or Mac-1 T209A (bottom) binding to fibrinogen (Fbg; A) and GP1bαN (B), and shown in wild-type Mac-1 I-domain binding to the GP1bαN mutants H218A (C, top) and E222A (C, bottom). RU, response unit.
A ligand surface of GPIbαN was generated through amine-coupling to a CM5 chip. To determine the integrity of the GPIbαN surface, a dilution series with the antibody Xia.B2 (emfret ANALYTICS) was conducted and analysis of sensorgrams resulted in a Kd of 34 nM, consistent with the anticipated high affinity for an antibody-antigen interaction, confirming the immobilized GPIbαN is forming an active surface. Sensorgrams were recorded using wild-type Mac-1 I-domain as the analyte and high-quality SPR curves were obtained (Figure 5B, top), such that association and dissociation profiles fitted well to a 1:1 Langmuir simultaneous binding model. A calculated Kd of 64 μM was in good agreement with the steady-state equilibrium Kd value of 121 μM.
Our model of the Mac-1 I:GPIbαN complex shows that the Mac-1 I-domain T209 is a pivotal residue at the binding interface not only by coordinating the MIDAS metal ion, but also in forming a hydrogen bond from the β-OH to the carboxylate of GPIbαN D222. The binding to GPIbαN of the Mac-1 T209 alanine variant (T209A) elicited neither a significant signal response nor a clear association binding curve (Figure 5B, bottom), which was similarly the case with wild-type Mac-1 with EDTA in excess (5 mM) to remove the metal ion from the MIDAS (data not shown). These experiments demonstrate that the bound Mg2+ ion in the MIDAS is critical to the interaction and confirm that the observed distal CSP effects in the NMR titration data arise from allosteric effects propagated away from the MIDAS site, rather than to a second ligand-binding site. In contrast with GPIbαN, at least 1 of these binding interactions is still evident from the SPR analysis with full-length fibrinogen and Mac-1 T209A, although the overall binding affinity is greatly reduced (Figure 5A, bottom). This is consistent with a primary binding site around the MIDAS surface, along with a second, low-affinity Mac-1 I-domain site.3
We further tested the model of the Mac-1 I:GPIbαN complex by generating recombinant mouse GP1bαN variants H218A and E222A, which were purified with a similar protocol and produced a similar yield to the wild-type protein (supplemental Figure 1A). SPR studies using the immobilized GPIbαN mutants with the wild-type Mac-1 I-domain as the analyte revealed that GP1bαN H218A and E222A substitutions led to a loss of SPR response, preventing the sensorgrams from being fitted using either kinetic or equilibrium binding models, indicating significant impairment of the interaction with the Mac-1 I-domain (Figure 5C).
Discussion
Interactions among innate immune cells, platelets, and plasma proteins have been implicated in pathways leading to thrombosis,10,27-29 and yet very few of these protein interactions have been fully described. To the best of our knowledge, we provide the first detailed description of the complex formed between the leukocyte integrin Mac-1 I-domain and the platelet receptor GPIbαN using a combination of NMR, crystallography, site-directed mutagenesis, and SPR binding studies. We establish the principle that the Mac-1 I-domain in the open conformation has a groove on the surface formed by the extended sidechains of F246 and R208 and other residues that can accommodate an α-helix appropriately positioned by an acidic side chain coordinating the MIDAS metal ion and the side chain hydroxyl of T209. We show that the GPIbαN receptor LRR C-terminal capping α-helix uses a key acidic residue (D222 in the human; E222 in the mouse ortholog) to coordinate to the vacant site in the metal ion coordination shell of the Mac-1 MIDAS Mg2+ ion.
The presence of the integrin-binding acidic residue at the end of an α-helix in GPIbαN is reminiscent of the crystal structure of the Mac-1 I-domain complex with iC3b, whereby the integrin MIDAS metal ion binding residue D1247 of iC3b is also placed at the end of an α-helix.4 A comparison of the Mac-1 I:iC3b and Mac-1 I:GPIbαN complexes reveals the MIDAS-coordinating α-helix is oriented differently (90° rotation) in each ligand relative to the Mac-1 I-domain surface. In addition, unlike GP1bαN, iC3b does not use the negatively charged patch (E244) on the surface of the Mac-1 I-domain for binding, but in common with GP1bαN it does use Mac-1 I-domain residue R208.4 These interactions suggest that the ability of the Mac-1 I-domain to bind a host of specific ligands involves a common binding surface and fits with the previously proposed mosaic model of Mac-1 ligand binding.30 By contrast, GPIbαN uses 3 spatially separated regions of the receptor to bind the structurally distinct ligands of the von Willebrand factor A1 domain24,26 (GPIbα LRRs and R-loop), thrombin25,31 (GPIbα anionic region), and Mac-1 (GPIbα C-terminal LRR cap).
The Mac-1 MIDAS interaction with an acidic residue from an α-helix of the ligand contrasts to the closely related integrin leukocyte function associated antigen-1 (LFA-1), which exploits a glutamate residue located within a β strand of the ligand ICAM-132 to bind the LFA-1 MIDAS α-subunit I-domain. A variety of studies have defined a large allosteric conformational shift between open and closed states of the leukocyte integrins Mac-1 and LFA-1 linked to the disposition of α-I-domain helix α7 and switching of the coordination shell within the MIDAS between D140 and T209.7,9 NMR studies have been previously reported for the related LFA-1 I-domain,33 but to date no NMR characterization has been applied to Mac-1. LFA-1 does not bind GPIbαN, and a series of LFA-1/Mac-1 chimeras have been used previously to localize the binding site for GPIbαN within the Mac-1 I-domain to specific residues. Many of these are consistent with the Mac-1:GP1bαN interface shown in Figure 4,22 but residues T213 and R216 are not part of the MIDAS face of the Mac-1 I-domain structure.10 The crystal structures of Mac-1 I-domain reveals the T213 side chain hydroxyl group is buried and hydrogen bonded by contact with the D248 side chain in both the open and closed conformation and thus is not present on the surface to interact with a ligand. Mac-1 residues T213 and R216 are very close to key MIDAS face residues R208 and F246 and thus may affect binding indirectly, or, alternatively, R216 could form part of a second binding site for GP1bαN.
Overall, we believe this is the first detailed description of the interface formed between an integrin receptor and an LRR protein revealing an elongated pocket in the GPIbαN LRR C-terminal capping region, which is key to the interaction. The Mac-1 integrin is not the only integrin to engage multiple ligands and, interestingly, the more distantly related integrin α2β1 binds to collagen19 as the principal ligand and has a well-characterized additional interaction with the LRR C-terminal capping region of the matrix protein chondroadherin.34 The binding site for the α2β1 integrin is localized to the chondroadherin sequence of residues LRRWLEAK,34 which resembles the FRRWLQDN sequence we report for GPIbα. Chondroadherin crystal structures reveal the LRRWLEAK sequence has an acidic residue that is surface exposed within the α-helix of the LRR capping region in a similar manner to GPIbαN. Previous authors also have speculated that the groove adjacent to the α-helix could be used by the integrin as described here.35,36 This structural scaffold describing an integrin-LRR protein interaction will facilitate the development of agents targeting Mac-1 or GPIbαN for the modulation of thrombotic and inflammatory disorders.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors acknowledge the Diamond Light Source for provision of synchrotron radiation in using the beamline I24 (Mac-1) and the European Synchrotron Radiation Facility (GPIbαN) and thank Phillip Williams for analysis of SPR data and Bernhard Neiswandt (emfret ANALYTICS) for providing the Xia.B2 antibody used in the SPR studies and Keith McCrae for the kininogen complementary DNA.
The work was supported in part by a British Heart Foundation grant RG/12/9/29775 and IG/16/1/32140 (J.E.), and by the School of Chemistry at the University of Nottingham and the EPSRC for studentship support (J.M.). The nuclear magnetic resonance facilities are supported by the School of Chemistry and were funded from the Nottingham HEFCE Capital Investment Fund allocation (2009).
Authorship
Contribution: J.A.L., M.S.S., and J.E. designed the research project; J.M. collected and assigned the nuclear magnetic resonance data with technical assistance from H.E.L.W., with nuclear magnetic resonance analysis of additional mutants by C.A.; cloning, protein expression, and isotopic labeling was carried out by J.M. with additional mutants produced by A.F.; X-ray analysis of mouse GP1bαN was carried out by M.S. and Mac-1 I-domain by R.N. and S.G.C.; surface plasmon resonance experiments were conducted by J.M., S.S.W., and A.D.M.; all authors discussed the results; and J.E., M.S.S., J.A.L., and J.M. contributed to the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.S.W. is School of Pharmacy, Keele University, Keele, United Kingdom.
Correspondence: Jonas Emsley, School of Pharmacy, Centre for Biomolecular Sciences, University of Nottingham, University Park, Nottingham NG7 2RD, United Kingdom; e-mail: jonas.emsley@nottingham.ac.uk; and Mark S. Searle, School of Chemistry, Centre for Biomolecular Sciences, University Park, University of Nottingham, Nottingham NG7 2RD, United Kingdom; e-mail: mark.searle@nottingham.ac.uk.