Key Points
FOXO1 mutations occur in 1/3 of sporadic and 1/2 endemic pediatric BL cases and cluster in and adjacent to the AKT recognition motif.
FOXO1 mutations occur in the major clone at initial diagnosis and are retained at relapse but are not associated with outcome.
Abstract
FOXO1 has an oncogenic role in adult germinal center–derived lymphomas, in which mutations, predominately within the AKT recognition motif, cause nuclear retention of FOXO1, resulting in increased cell proliferation. To determine the prevalence and distribution of FOXO1 mutations in pediatric Burkitt lymphoma (BL), we sequenced a large number of sporadic and endemic BL patient samples. We report a high frequency of FOXO1 mutations in both sporadic and endemic BL at diagnosis, occurring in 23/78 (29%) and 48/89 (54%) samples, respectively, as well as 8/16 (50%) cases at relapse. Mutations of T24 were the most common in sporadic BL but were rare in endemic cases, in which mutations of residue S22, also within the AKT recognition motif, were the most frequent. FOXO1 mutations were almost always present in the major tumor cell clone but were not associated with outcome. Analysis of other recurrent mutations reported in BL revealed that FOXO1 mutations were associated with mutations of DDX3X and ARID1A, but not MYC, TCF3/ID3, or members of the phosphatidylinositol 3-kinase signaling pathway. We further show common nuclear retention of the FOXO1 protein, irrespective of mutation status, suggesting alternative unknown mechanisms for maintaining FOXO1 transcriptional activity in BL. CRISPR/Cas9 knockout of FOXO1 in an endemic cell line produced a significant decrease in cell proliferation, supporting an oncogenic role for FOXO1 in endemic BL. Thus, FOXO1 is frequently mutated in both sporadic and endemic BL and may offer a potential therapeutic target for pediatric BL patients worldwide.
Introduction
The forkhead box subtype O (FOXO) family of transcription factors have pleiotropic roles in development and immunity and context-dependent tumor suppressor and oncogenic capacity.1-3 During the germinal center reaction, FOXO1 controls B-cell trafficking through cycles of clonal expansion and affinity maturation.4,5 Within the normal germinal center, nuclear FOXO1 directs a gene expression program in the dark zone, in which B cells proliferate and undergo immunoglobulin somatic hypermutation, but FOXO1 activity is downregulated by phosphatidylinositol 3-kinase (PI3K) signaling in the light zone, where cells undergo affinity selection and further differentiation. Here, B-cell receptor (BCR) signaling through the PI3K cascade causes AKT-dependent phosphorylation of the T24 residue of FOXO1, resulting in sequestration by 14-3-3 in the cytoplasm, nuclear exclusion, and loss of transcriptional activity.6,7 Thus, FOXO1 and BCR/PI3K work in opposition as regulators of the germinal center reaction.
Given the critical role of BCR/PI3K signaling in the pathogenesis of germinal center–derived lymphomas8 such as Burkitt lymphoma (BL) and diffuse large B-cell lymphomas (DLBCL),9,10 the occurrence of FOXO1 mutations in these malignancies is of great interest. FOXO1 mutations have been identified in up to 11% of adult DLBCL,11-18 and FOXO1 has been identified as a driver gene in DLBCL cell lines.15 DLBCL with mutations of the M1 residue, resulting in loss of the AKT recognition motif, or within this motif itself, have aberrant nuclear localization of FOXO1 and are associated with a poorer prognosis.17 Activating FOXO1 mutations have also been identified in tumors from a murine MYC/PI3K-driven model of BL and in human BL cell lines.19 These models demonstrate the role of T24 mutations in uncoupling FOXO1 from BCR/PI3K, driving nuclear localization of FOXO1 protein with subsequent pro-proliferative and pro-survival effects. However, to date, next-generation sequencing (NGS) based studies have identified FOXO1 mutations in only a small proportion of BL patient samples,20-24 raising the question of how FOXO1 functions in the setting of BCR/PI3K signaling in BL.
We report a comprehensive analysis of FOXO1 mutations in the largest cohort of pediatric sporadic and endemic BL patient samples sequenced to date. We show a high frequency of FOXO1 mutations in pediatric sporadic and endemic BL, revealing different mutation hotspots within the AKT recognition motif in the 2 BL subtypes. We show nuclear localization of FOXO1 in patient samples and cell lines and provide functional knockout data supporting an important role for FOXO1 as an oncogenic factor in the pathogenesis of BL.
Methods
Patient samples and cell lines
Sporadic BL samples from UK hospitals were deposited in the Children’s Cancer and Leukaemia Group (CCLG) Tissue Bank between 1995 and 2014. Project approval was obtained from the CCLG Biological Studies Steering Group (2012 BS 08). CCLG samples were stored either frozen or following formalin fixation and paraffin embedding. Endemic BL samples were collected at Queen Elizabeth Central Hospital, Blantyre, Malawi, between 2009 and 2016, with ethical approval from the College of Medicine Research and Ethics Committee (Malawi). Surplus material from fine-needle aspirates was fixed in ThinPrep PreservCyt Solution (Hologic, Manchester, United Kingdom) and stored at ambient temperature. For both sporadic and endemic BL, diagnoses were confirmed by pathology review according to World Health Organization criteria using immunocytochemistry (including CD20, CD3, CD10, BCL2, BCL6, and Ki67), in situ hybridization for Epstein-Barr virus–encoded small RNA and fluorescence in situ hybridization for MYC translocations.25 Sporadic BL patients were treated according to French-American-British/Lymphome Mains de Burkitt–based protocols,26-28 and endemic BL patients were treated with low-intensity therapy based on a published 28-day regimen29,30 (Table 1).
Clinical and molecular characteristics of BL cohorts
| . | Sporadic BL . | Endemic BL . |
|---|---|---|
| Total cases, n | 78 | 89 |
| Median age at diagnosis (range), y | 8 (0-19) | 7 (2-16) |
| Sex, n (%) | ||
| Male | 61 (78.2) | 57 (64.0) |
| U | 5 (6.4) | 1 (1.1) |
| Tumor stage, n (%) | ||
| High (3 or 4) | 30 (38.5) | 58 (65.2) |
| U | 38 (48.7) | 8 (9.0) |
| CSF involvement, n (%) | ||
| Y | 1 (1.3) | 10 (11.2) |
| U | 28 (36.0) | 25 (28.1) |
| BM involvement, n (%) | ||
| Y | 18 (23.1) | 8 (9.0) |
| U | 17 (21.8) | 33 (37.1) |
| CNS involvement, n (%) | ||
| Y | 4 (5.1) | ND |
| U | 23 (29.5) | |
| EBV positive, n (%) | ||
| Y | 4 (5.1) | 76 (85.4) |
| U | 22 (28.2) | 13 (14.6) |
| MYC translocation, n (%) | ||
| Y | 73 (93.6) | 89 (100) |
| U | 1 (1.3) | 0 (0) |
| Treatment, n (%) | ||
| FAB/LMB 96 Protocol | 63 (81) | — |
| Group A | 1 | — |
| Group B | 44 | — |
| Group C | 14 | — |
| Group unknown | 4 | — |
| Malawi 28 d protocol* | — | 78 (88) |
| With doxorubicin | — | 60 |
| Reduced protocol | — | 18 |
| No treatment | 1 (1) | 5 (6) |
| U | 14 (18) | 6 (7) |
| Follow-up | ||
| Number of cases with follow-up data (%) | 71 (91.0) | 71 (79.8) |
| Disease progression | ||
| Relapse/refractory disease, n | 10 | 22 |
| Median time to relapse (range), mo | 5.5 (2.8-6.7) | 5.1 (1.6-10.9) |
| DFS | ||
| Progression or death from lymphoma, n | 12 | 30 |
| Median time to event (range), mo | 4.83 (0.9-6.7) | 3.92 (0.03-10.9) |
| OS | ||
| Dead, n | 12 | 31 |
| Median time to death (range), mo | 4.42 (0.1-11.1) | 4.73 (0.03-14.6) |
| . | Sporadic BL . | Endemic BL . |
|---|---|---|
| Total cases, n | 78 | 89 |
| Median age at diagnosis (range), y | 8 (0-19) | 7 (2-16) |
| Sex, n (%) | ||
| Male | 61 (78.2) | 57 (64.0) |
| U | 5 (6.4) | 1 (1.1) |
| Tumor stage, n (%) | ||
| High (3 or 4) | 30 (38.5) | 58 (65.2) |
| U | 38 (48.7) | 8 (9.0) |
| CSF involvement, n (%) | ||
| Y | 1 (1.3) | 10 (11.2) |
| U | 28 (36.0) | 25 (28.1) |
| BM involvement, n (%) | ||
| Y | 18 (23.1) | 8 (9.0) |
| U | 17 (21.8) | 33 (37.1) |
| CNS involvement, n (%) | ||
| Y | 4 (5.1) | ND |
| U | 23 (29.5) | |
| EBV positive, n (%) | ||
| Y | 4 (5.1) | 76 (85.4) |
| U | 22 (28.2) | 13 (14.6) |
| MYC translocation, n (%) | ||
| Y | 73 (93.6) | 89 (100) |
| U | 1 (1.3) | 0 (0) |
| Treatment, n (%) | ||
| FAB/LMB 96 Protocol | 63 (81) | — |
| Group A | 1 | — |
| Group B | 44 | — |
| Group C | 14 | — |
| Group unknown | 4 | — |
| Malawi 28 d protocol* | — | 78 (88) |
| With doxorubicin | — | 60 |
| Reduced protocol | — | 18 |
| No treatment | 1 (1) | 5 (6) |
| U | 14 (18) | 6 (7) |
| Follow-up | ||
| Number of cases with follow-up data (%) | 71 (91.0) | 71 (79.8) |
| Disease progression | ||
| Relapse/refractory disease, n | 10 | 22 |
| Median time to relapse (range), mo | 5.5 (2.8-6.7) | 5.1 (1.6-10.9) |
| DFS | ||
| Progression or death from lymphoma, n | 12 | 30 |
| Median time to event (range), mo | 4.83 (0.9-6.7) | 3.92 (0.03-10.9) |
| OS | ||
| Dead, n | 12 | 31 |
| Median time to death (range), mo | 4.42 (0.1-11.1) | 4.73 (0.03-14.6) |
BM, bone marrow; CNS, central nervous system; CSF, cerebrospinal fluid; EBV, Epstein-Barr virus; FAB-LMB, French-American-British/Lymphome Mains de Burkitt; U, unknown; Y, yes.
Endemic BL patients were treated with low-intensity 28-d regimen,29,30 receiving cyclophosphamide (cyclo), vincristine (vcr), prednisolone (pred), and doxorubicin (dox). Eighteen endemic BL patients received a reduced protocol (4 had cyclo only, 6 cyclo+vcr, and 8 cyclo+vcr+pred).
To generate patient-derived xenograft models, diagnostic patient samples were retrieved from the Newcastle Biobank and transplanted into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice in accordance with the Biobank’s ethical approval (NRES Committee, 16/NE/0002) and the Home Office project license PPL 60/4552. Resultant tumors were harvested and mechanically disaggregated to provide single-cell suspensions. BL cell lines were purchased from DSMZ (Germany) and cultured in Gibco RPMI 1640 medium with 10% fetal bovine serum (Fisher Scientific, Loughborough, United Kingdom).
Sanger sequencing
DNA and RNA were extracted using Qiagen Allprep Kit. Primers were designed to amplify the coding region of the FOXO1 transcript ENST00000379561 (supplemental Table 1). The expression of mutant alleles in cell lines was confirmed by reverse transcriptase polymerase chain reaction (PCR) using the High Capacity RNA-to-cDNA Kit (Fisher Scientific). DNA was amplified using the Repli-g Kits (Qiagen, Manchester, United Kingdom) for cases with small samples and low DNA yields. PCR products were amplified using the AmpliTaq Gold DNA Polymerase kit (Fisher Scientific) and amplification of the GC-rich exon 1 was enhanced by addition of 5% v/v dimethyl sulfoxide and 0.8M betaine solution (Sigma-Aldrich, Gillingham, United Kingdom).17 PCR products were amplified using the SureCycler 8800 (Agilent, Cheshire, United Kingdom) and then purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced by Eurofins Genomics (Ebersberg, Germany).
ABI format files were analyzed using GeneScreen and visualized using FinchTV software (Geospiza).31 Heterozygous mutation calls required a >0.2 peak height ratio and all mutated bases had a quality score >0. Each ABI chromatogram was manually checked to ensure the absence of background noise, “dye blobs,” low signal, or discordant mutation calls between forward and reverse reads. Any conflicts or poor-quality reads were resequenced to remove potential false positives. The high GC content of exon 1 proved challenging when designing nested PCR primers for both the forward and reverse for 2 regions; therefore, 3 mutations in endemic (S205T, S206L, A207P) and 2 in sporadic BL (both D82N) could be identified in 1 direction only. In 14 sporadic and 8 endemic BL cases, the FOXO1 mutations were shown to be somatic nonsynonymous mutations by sequencing matched constitutional DNA. Where constitutional DNA was unavailable, mutations were analyzed against the Catalogue of Somatic Mutations in Cancer, the Single Nucleotide Polymorphism database, and the Exome Aggregation Consortium database to remove germline variants.
Next-generation sequencing and mutation data analysis
Whole-exome sequencing (WES) data were generated for 74 sporadic BL samples using Illumina Nextera Exome enrichment by the Newcastle University Genomics Core Facility. Because of the low yields and poor quality of DNA from fine-needle aspirates, WES data could not be generated for the endemic BL cohort. Data were analyzed using Genome Analysis Toolkit32,33 and aligned using BWA-MEM. Aligned SAM files were sorted, converted to BAM, indexed, and PCR duplicates marked using Picard. Variants were called using MuTect.34 Matched constitutional DNA was used to call variants where available. For cases with no matched DNA, a pool of 33 constitutional DNAs from B-cell non-Hodgkin lymphoma patients with no other underlying conditions was used as the reference. Variants were annotated with gene and gene function data from Ensembl’s Variant Effect Predictor. Data were visualized in Integrative Genomics Viewer.35 Comparative genomic analysis to determine the conservation of residues within the AKT recognition motif was performed. FOXO1 protein sequences for Homo sapiens and 10 other species were downloaded from the Uniprot database and aligned using MUSCLE software.36 Alignments were visualized with Jalview software.37
FOXO1 mutations in BL samples were identified in the supplementary files of 5 published studies.20-24 To investigate the coverage of the FOXO1 gene, raw FASTQ files were downloaded from dbGAP and analyzed as described above. FOXO1 mutations and sequence coverage were visualized in Integrative Genomics Viewer.
Data are available from the European Genome-phenome Archive accession number EGAS00001003719.
Survival analysis
Estimates of overall survival (OS) and disease-free survival (DFS) were calculated and compared using Kaplan-Meier methods, log-rank tests, and univariate Cox regression models. For both sporadic and endemic BL, OS was defined as the time from diagnosis to death from any cause, with censoring at the date of last contact. DFS was calculated from the date of diagnosis to relapse or death from lymphoma, censoring deaths from other causes. Factors present were considered to be significant when P < .05. All variables conformed to the proportional hazards assumption. Other comparisons were performed using the Fisher’s exact test. Analyses were performed using R Bioconductor packages “survival” for univariate analysis and “survminer” for visualization of Kaplan-Meier survival curves, using modified in-house R scripts.
FOXO1 knockout by CRISPR/Cas9 gene editing
CRISPR/Cas9 single guide sequences specifically targeting FOXO1 (sgFOXO1) were designed using the CHOPCHOPv2 Web tool and cloned into the lentiCRISPRv2 plasmid. The plasmid was a kind gift from Fen Zhang (Addgene plasmid #52961; http://n2t.net/addgene:52961; RRID:Addgene_52961).38 The nontargeting control sequence used was 5′-TGAGGATCATGTCGAGCGCC obtained from the GeCKOv2 library (Addgene). Lentiviral particles were produced in 293T cells and concentrated by ultracentrifugation. BL cell lines were transduced with sgFOXO1 lentiviruses or nontargeting control and puromycin (Santa Cruz Biotechnology, Dallas, TX) was added on day 4, using the minimal toxic dose per cell line to select transduced cells. Lentiviral transduction efficiency was established on day 6 by measuring the percentage of viable cells selected by puromycin relative to nonselected control cells. Viral titers with transduction efficiencies of <40% were used to avoid excessive multiple viral integrations. Single colonies for Jiyoye and Ramos were selected by methylcellulose colony formation assay and limiting dilution assay, respectively. The effect of FOXO1 gene editing on cell proliferation was assessed by Trypan Blue viable cell counting over a 7-day time course, and for apoptosis by flow cytometry (BD FACSCalibur) using Annexin V/7-AAD (BD Biosciences). For each cell line, 3 independent experiments were performed for both the bulk and clonal populations. The GraphPad Prism 6 software (GraphPad Software, USA) was used for 2-tailed unpaired Student t test, as well as plotting histograms and scatter plots. Statistical significance was considered as P < .05. Total protein from BL cells was extracted using Pierce RIPA buffer (Fisher Scientific) and cytoplasmic and nuclear protein was extracted with the NE-PER Nuclear and Cytoplasmic Extraction Kit (Fisher Scientific). Protein expression was determined by western blotting probed for FOXO1 (Cell Signaling, 2880S), PARP (9542S; Cell Signaling) or α-Tubulin (T6199; Sigma-Aldrich) with Amersham ECL detection (GE Healthcare Life Sciences, Buckinghamshire, United Kingdom).
Results
Clinical and molecular characteristics of the sporadic and endemic BL cohorts
Diagnostic samples taken at first presentation were obtained from 78 patients with sporadic BL and 89 patients with endemic BL. The clinical and molecular characteristics of these cohorts are summarized in Table 1. The median follow-up for sporadic BL was 49.4 (0.1-228.6) months and for endemic BL was 12.1 (0.03-45.6) months. Survival estimates at 12 months for OS and DFS were 83.1% (95% confidence interval [CI]. 74.8-92.3) and 82.4% (95% CI, 73.8-92.0), respectively, for the sporadic BL and 70.5% (95% CI, 60.0-82.7) and 54.4% (95% CI, 43.6-68.0), respectively, for endemic BL. The median survival time was not reached in either cohort. For those sporadic and endemic BL patients who had progressive disease (relapse or primary refractory disease), the median time from initial diagnosis to progression was 5.5 and 5.1 months, respectively. We found MYC rearrangements in 73 of 78 (94%) sporadic BL. Sixty-nine sporadic BL cases had an IG-MYC translocation, 4 had a MYC translocation but the partner was unconfirmed, 1 failed fluorescence in situ hybridization, and 4 had no detectable MYC translocation. All endemic BL cases (89/89; 100%) carried an IG-MYC rearrangement. Tumor EBV status was determined by Epstein-Barr virus–encoded small RNA in situ hybridization. EBV status was available for 56 and 76 sporadic and endemic BL samples, respectively, with 4/56 (7.1%) sporadic and all 76/76 (100%) endemic BL cases EBV positive.
FOXO1 mutations occur at high frequency in sporadic and endemic BL
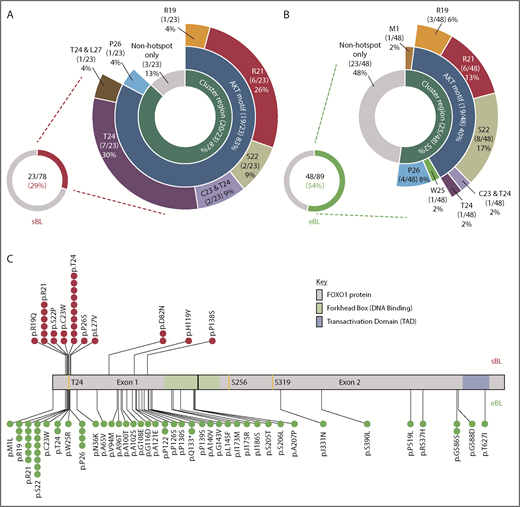
Using Sanger sequencing, we identified 27 FOXO1 mutations in 23/78 (29%) sporadic BL samples, and 59 mutations in 48/89 (54%) endemic BL samples taken at first diagnosis (Figure 1A-B; supplemental Tables 2 and 3). Of the 86 mutations, 83 were missense mutations with only 1 M1 mutation and 2 Q133* nonsense mutations detected. The frequency of mutations contrasts strikingly with the substantially lower frequencies previously reported using NGS approaches: 4/86 (5%) sporadic BL and 2/48 (4%) endemic BL (supplemental Figure 1; supplemental Table 4).20-24 To confirm the observed FOXO1 mutational status, we analyzed WES data for 74/78 sporadic samples. This included 21 of the 23 mutated cases, in which a total of 25 FOXO1 mutations had been identified by Sanger sequencing. WES confirmed 21/25 (84%) of those mutations (examples shown in supplemental Figure 2). WES analysis highlighted poor coverage of the GC-rich exon 1 compared with exon 2 and all 4 mutations (D82N in 2 samples, H119Y and P138S) not confirmed by WES were located within a region of exon 1 with no or virtually no read coverage (supplemental Figure 3). No additional FOXO1 mutations were detected in any of the 74 cases analyzed by WES.
Overview of FOXO1 mutations in pediatric sporadic and endemic BL patients. The distribution of mutations in patient samples identified in the evolutionarily conserved cluster region (M1, R19-L27), including the AKT motif (R19-T24), are shown for the 23 of 78 FOXO1 mutated sporadic BL (sBL; red) (A) and the 48 of 89 FOXO1-mutated endemic BL (eBL; green) (B) samples, taken at initial diagnosis. Fourteen patients had multiple FOXO1 mutations: in 4 cases, 2 mutations were both located within the cluster region (as indicated), 3 had an S22 mutation with a second mutation located outside the cluster region and the remaining 7 cases had 2 or more mutations located outside the cluster region only. (C) Mutation lolliplot showing the positions of all mutations detected in the FOXO1 protein relative to the phosphorylation sites T24, S256, and S319 (yellow) and protein domains. Mutations in the sBL cohort are shown at top in red and in the eBL cohort at bottom in green.
Overview of FOXO1 mutations in pediatric sporadic and endemic BL patients. The distribution of mutations in patient samples identified in the evolutionarily conserved cluster region (M1, R19-L27), including the AKT motif (R19-T24), are shown for the 23 of 78 FOXO1 mutated sporadic BL (sBL; red) (A) and the 48 of 89 FOXO1-mutated endemic BL (eBL; green) (B) samples, taken at initial diagnosis. Fourteen patients had multiple FOXO1 mutations: in 4 cases, 2 mutations were both located within the cluster region (as indicated), 3 had an S22 mutation with a second mutation located outside the cluster region and the remaining 7 cases had 2 or more mutations located outside the cluster region only. (C) Mutation lolliplot showing the positions of all mutations detected in the FOXO1 protein relative to the phosphorylation sites T24, S256, and S319 (yellow) and protein domains. Mutations in the sBL cohort are shown at top in red and in the eBL cohort at bottom in green.
Sporadic and endemic BL have different mutation hotspots within the AKT recognition motif
The majority of recurrent FOXO1 mutations clustered within or close to the AKT recognition motif (RxRxxS/T) at amino acid residues 19 through 24 (Figure 1A-C), implying a role in aberrant nuclear localization of FOXO1. Overall, 23 and 26 FOXO1 mutations were identified involving the evolutionarily conserved cluster region between residues R19 and L27 and the M1 residue in 20 of 78 sporadic and 25 of 89 endemic BL samples obtained at diagnosis, respectively (supplemental Figure 4). Despite their similar combined frequencies, the distribution of mutations within this cluster region differed between subtypes. Mutations of the T24 phosphorylation site were the most common mutation in sporadic BL, representing 10/23 (37%) mutations within the cluster region. Strikingly, only 2/26 (8%) mutations in the cluster region of endemic BL cases involved T24, whereas mutations of S22 were instead the most common, being seen in 8/26 (31%) samples. Mutations were also identified distal to the AKT recognition motif. Most commonly these were nonrecurrent mutations in endemic BL, but we also identified previously unreported recurrent mutations at P122, Q133, and G586 in endemic BL and D82 in sporadic BL. These mutations were predominantly located in exon 1, but most were outside the DNA binding domain and their functional relevance remains unclear.
Mutations in ARID1A and DDX3X are associated with FOXO1 mutations
We next sought to determine, in our sporadic BL cohort, the relationship between FOXO1 mutations and additional mutations previously reported to be recurrent in BL. In keeping with previous reports, the most commonly mutated genes were MYC, ID3, and TP53 occurring in 47/74 (64%), 44/74 (59%), and 37/74 (50%) cases, respectively (Figure 2). Among the mutations examined, mutations in 2 genes were associated with FOXO1 mutations: DDX3X (present in 8/21 FOXO1 mutated vs 7/53 FOXO1 nonmutated cases, P = .025) and ARID1A (present in 9/21 FOXO1 mutated vs 8/53 FOXO1 nonmutated cases, P = .016). Six of the 15 DDX3X mutations were nonsense mutations, the remaining 9 were missense mutations, and all but 1 were located in either the DEAD box helicase domain or the Helicase C terminus supplemental Figure 5A). Eighteen ARID1A mutations were detected in 17 cases, including 3 frameshift mutations, 10 nonsense mutations, and 5 missense mutations (supplemental Figure 5B). These mutations were distributed equally throughout the ARID1A protein.
Survival analysis. Kaplan-Meier plots showing association of FOXO1 mutations with overall survival in sporadic BL (A) and endemic BL (B) and DFS in sporadic BL (C) and endemic BL (D).
Survival analysis. Kaplan-Meier plots showing association of FOXO1 mutations with overall survival in sporadic BL (A) and endemic BL (B) and DFS in sporadic BL (C) and endemic BL (D).
To determine whether FOXO1 mutations were present in the dominant tumor cell clone we compared the variant allele frequencies of FOXO1 point mutations with those of other mutations present in the same samples (supplemental Table 5). Eighteen sporadic samples contained FOXO1 mutations detected by WES, and in almost all of these (16/18; 89%), the FOXO1 mutations were present in the major clone. In only 2 cases, the variant allele frequencies of the FOXO1 mutations were substantially lower than those of other mutations, suggesting that they were present within a sub-clonal population.
FOXO1 mutations are not associated with outcome in BL subtypes but are retained at relapse
Given a reported association between FOXO1 mutations and a poor outcome in DLBCL,17 we assessed the association between FOXO1 mutation status and prognosis in both the sporadic and endemic BL cohorts. We found that neither OS nor DFS was associated with FOXO1 mutation status in either BL subtype (Figure 3). We also investigated the prognostic significance of mutations within the AKT binding motif (R19-T24) and the extended evolutionarily conserved cluster region (M1 and R19-L27), in which mutations have been predicted to result in persistent FOXO1 transcriptional activity. However, there was no association with outcome in our cohorts (supplemental Table 6).
Overview of somatic mutations identified in the pediatric sporadic BL cohort. Mutations affecting genes previously identified as commonly mutated in BL are shown. Each column represents a BL sample, ordered by the presence or absence of FOXO1 mutations. EBV status is indicated as positive or negative. Mutations are colored-coded by type. The prevalence of different mutations in FOXO1 mutated (blue) and nonmutated (orange) cases is shown in the right-hand barcharts and genes significantly more commonly mutated in FOXO1 mutated cases are indicated by an asterisk (Fisher’s exact test). EBV, Epstein-Barr virus; Fs, frameshift.
Overview of somatic mutations identified in the pediatric sporadic BL cohort. Mutations affecting genes previously identified as commonly mutated in BL are shown. Each column represents a BL sample, ordered by the presence or absence of FOXO1 mutations. EBV status is indicated as positive or negative. Mutations are colored-coded by type. The prevalence of different mutations in FOXO1 mutated (blue) and nonmutated (orange) cases is shown in the right-hand barcharts and genes significantly more commonly mutated in FOXO1 mutated cases are indicated by an asterisk (Fisher’s exact test). EBV, Epstein-Barr virus; Fs, frameshift.
To further determine whether FOXO1 mutations might be associated with disease progression, we sequenced FOXO1 in 5 sporadic and 11 endemic BL samples obtained at the time of disease relapse. FOXO1 mutations were detected in relapse samples at frequencies similar to those seen at presentation: 2/5 (40%) sporadic and 6/11 (55%) endemic BL relapse samples (supplemental Figure 6). Overall, 9 mutations were detected, of which 6 were located within or juxtaposed to the AKT recognition motif. Four matched diagnostic and relapse samples were concordant for FOXO1 mutations: 2 patients, 1 sporadic and 1 endemic BL, had FOXO1 mutations (R21H and P26L) and 2 were unmutated at both initial diagnosis and relapse. WES data were not available for the 2 mutated cases, so quantitative analysis of allele frequency could not be performed.
CRISPR/Cas9 knockout supports an oncogenic role of FOXO1 in endemic BL
The demonstration of FOXO1 mutations in a high proportion of BL samples, the presence of these mutations in the dominant tumor cell clone in most cases, and the clustering of mutations in or around the AKT recognition motif suggest a role for FOXO1 in the pathobiology of BL. We screened 7 cell lines and 4 patient-derived xenograft samples for FOXO1 mutations and identified 7 mutations in 3 lines and 2 xenografts, including S22P in the endemic cell line Jiyoye and D82N in the sporadic BL cell line Ramos (Figure 4A). Both of these mutations were recurrent in the patient cohorts (Figure 1). The expression of the mutant alleles carrying S22P and S37W in Jiyoye, and D82N in Ramos was confirmed by Sanger sequencing. Interestingly, all patient-derived xenograft samples and cell lines showed strong nuclear retention of FOXO1, irrespective of mutation status (Figure 4B).
Investigation of FOXO1 function in endemic BL cells using CRISPR/Cas9 gene editing. (A) Summary of FOXO1 mutations detected in 7 human BL cell lines and 4 patient-derived xenografts. (B) Western blot showing FOXO1 protein expression and localization in BL cell lines and 4 patient-derived xenografts. PARP and α-tubulin were used as loading controls for the nuclear (N) and cytoplasmic (C) fractions, respectively. CRISPR/Cas9 gene editing in the Jiyoye cell line, analyzing bulk populations (C) and individual clones (D). Left panels show FOXO1 protein expression by western blot. Right panels show the effect on cell proliferation determined by Trypan Blue cell counting over a 7-day time course. (C) The mean and the standard deviation of 3 independent experiments are presented. (D) Each dot in the box-whisker plot (10th-90th percentile) represents the mean of 3 independent experiments performed with an individual clone. *P < .05, **P < .01, ***P < .001. NTC, nontargeting controls
Investigation of FOXO1 function in endemic BL cells using CRISPR/Cas9 gene editing. (A) Summary of FOXO1 mutations detected in 7 human BL cell lines and 4 patient-derived xenografts. (B) Western blot showing FOXO1 protein expression and localization in BL cell lines and 4 patient-derived xenografts. PARP and α-tubulin were used as loading controls for the nuclear (N) and cytoplasmic (C) fractions, respectively. CRISPR/Cas9 gene editing in the Jiyoye cell line, analyzing bulk populations (C) and individual clones (D). Left panels show FOXO1 protein expression by western blot. Right panels show the effect on cell proliferation determined by Trypan Blue cell counting over a 7-day time course. (C) The mean and the standard deviation of 3 independent experiments are presented. (D) Each dot in the box-whisker plot (10th-90th percentile) represents the mean of 3 independent experiments performed with an individual clone. *P < .05, **P < .01, ***P < .001. NTC, nontargeting controls
Using CRISPR/Cas9 gene editing, we investigated the effect of FOXO1 knockout in Jiyoye (S22P, S37W) and Ramos (D82N) cell lines using 3 guide sequences (supplemental Figure 7). In Jiyoye, FOXO1 editing with each of 3 single-guide RNAs caused almost complete loss of FOXO1 protein expression in bulk cell cultures and a significant decrease in cell proliferation compared with controls (Figure 4C). The decrease in cell proliferation was confirmed by the further investigation of multiple low-expressing, single-cell clones that also showed no significant increase in apoptosis, albeit this difference was not significant (Figure 4D; supplemental Figure 8). In contrast, a decrease in cell proliferation in the Ramos line, was only observed in the bulk population edited by sg3 and could not be confirmed by analysis of individual clones (supplemental Figure 9).
Discussion
This study, utilizing a large cohort of pediatric BL patient samples, reveals that FOXO1 mutations occur in both sporadic and endemic BL at substantially higher frequency than previously reported.20-24 This is likely from the different technical approaches used because coverage of FOXO1 on many NGS platforms is poor.17,39 We observed that the most frequent FOXO1 mutations were located within or immediately adjacent to the AKT recognition motif (RxRxxS/T), as has previously been reported in DLBCL.17 However, we also show for the first time that the hotspot mutations within the AKT recognition motif differ between BL subtypes, with the T24 residue most commonly affected in sporadic cases and S22 in endemic cases. In contrast to DLBCL, we identified only a single sporadic BL case with an M1 mutation, predicted to remove the AKT recognition motif by forcing use of an alternate translation start site. Furthermore, we identified mutations outside the AKT motif, predominantly in endemic BL, but in contrast to DLBCL,17 these did not cluster within the DNA-binding domain. The functional significance of these mutations, and whether the differences observed between sporadic and endemic BL represent differences in the regulation of FOXO1, or different mutational pressures or processes, remain to be explored.
In almost all cases, the FOXO1 mutations were present within the major tumor cell clone at initial diagnosis, supporting an important biological role for mutant FOXO1 in the development of BL. In concordance with our data, Grande et al have recently identified FOXO1 missense mutations in both endemic and sporadic BL in a high proportion of cases irrespective of tumor EBV status.40 FOXO1 mutations in DLBCL have been reported to be associated with a poor prognosis and to be acquired or expanded at relapse.17,39 In our study, we demonstrated persistence of FOXO1 mutation in paired presentation/relapse sample, consistent with a role throughout the disease course, but there was no correlation between FOXO1 mutations and clinical outcome in either sporadic or endemic BL. This could represent differences in the biology or treatment of DLBCL and BL, but the prognostic effect in DLBCL has also more recently been challenged.18
We identified 2 genes that were more frequently mutated in cases harboring FOXO1 mutations: the ATP-dependent RNA helicase DDX3X (DEAD-Box Helicase 3 X-Linked) and ARID1A (AT-rich interaction domain 1A), a member of the SWI/SNF chromatin-remodeling complex. DDX3X and ARID1A mutations are commonly found in BL and are frequently silencing events, such as nonsense and frameshift mutations. The functional relevance of the FOXO1-DDX3X/ARID1A association is, however, currently not clear. Notably, it has previously been reported that mutations in the SWI/SNF family members ARID1A and SMARCA4 are mutually exclusive.22 We did not observe this in our study, with 4 cases harboring mutations in both genes, suggesting that dysregulation of the SWI/SNF complex is more complicated in BL than initially proposed.
BL cells are believed to be dependent on signaling through the PI3K pathway.8,9 In physiological states, PI3K signaling drives AKT-dependent phosphorylation of FOXO1 at 3 specific sites (T24, S256, and S3196,17 ), resulting in binding to 14-3-3 and sequestration of FOXO1 in the cytoplasm.6,7 In apparent contradiction to this, we found abundant FOXO1 in the nucleus of both human BL cell lines and patient-derived xenograft samples, consistent with transcriptional activity. Mutations affecting the N-terminal AKT recognition motif, including those detected in this study, have been shown to prevent FOXO1 cytoplasmic localization,17 and indeed Kabrani et al have very recently demonstrated that T24 mutation drives nuclear retention of FOXO1 and thereby cell proliferation and survival in murine PI3K-driven, BL-like tumors carrying acquired FOXO1 mutations.19 In our study, FOXO1 knockout resulted in a significant decrease in cell proliferation in an endemic BL cell line, supporting an oncogenic function for FOXO1 in human BL as well. Thus, FOXO1 mutation likely represents 1 mechanism by which active PI3K signaling and FOXO1 activity can cooccur in BL cells. However, further complexities must exist in the control of FOXO1 in BL as nuclear localization is seen even in the absence of AKT recognition motif mutations. This lack of association between FOXO1 mutation status and FOXO1 nuclear localization may explain the lack of association with clinical outcome.
In conclusion, we have identified a previously undiscovered high frequency of FOXO1 mutations in pediatric sporadic and endemic BL. Together with previous findings that FOXO1 is a driver of in DLBCL15,17 and murine BL-like tumors19 and may mediate therapeutic resistance,41 our data further identify the transcription factor FOXO1 as an important factor in pathophysiology of several germinal center–derived B-cell lymphomas. Further exploration of the mechanisms uncoupling FOXO1 from PI3K signaling will aid understanding of BL pathogenesis. With small molecule inhibitors in development,42,43 FOXO1 may represent an emerging therapeutic target for a substantial proportion of pediatric BL patients at initial diagnosis and for the currently hard-to-treat children with relapsed disease in both high- and low-income countries.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank all the patients, clinicians, and staff who participated in the collection of material and clinical data at the Queen Elizabeth Central Hospital, Blantyre, and the Children’s Cancer and Leukaemia Group (CCLG) Tissue Bank.
This study was supported by a Bloodwise (formerly Leukaemia and Lymphoma Research) Senior Bennett Fellowship (#12005) (V.R.), the Newcastle upon Tyne Hospitals NHS Charity, Newcastle Promenaders Against Cancer (NEPAC), the JGW Patterson Foundation, the Little Princess Trust, the North of England Children’s Cancer Research Fund (NECCR), the Good Will Cause, and the MRC/EPSRC Newcastle Molecular Pathology Node. The CCLG Tissue Bank is funded by Cancer Research UK.
Authorship
Contribution: V.R. conceived the study, secured funding, and finalized the manuscript with editorial assistance from all authors; V.R., S. Bomken, and C.M.B. directed the research, designed the study, and wrote the manuscript; V.R. and P.Z. coordinated and participated in data collection, analysis, and interpretation; P.Z., A.E.B., S.V.N., I.L., F.R.A., U.T.O., C.B., L.C., A.W., A.H., and H.O. did laboratory experiments and analysis; V.R., A.M.N., and M.Z. performed statistical analysis; G.C., E.M., S. Bailey, V.R., C.M.B., and S. Bomken gathered samples and patient data and provided clinical interpretation; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vikki Rand, Wolfson Childhood Cancer Research Centre, Northern Institute for Cancer Research, Newcastle upon Tyne NE1 7RU, United Kingdom; e-mail: vikki.rand@newcastle.ac.uk.