Hematopoietic stem cells differentiate into all types of blood cells, including peripheral tissue-resident mast cells. The early mast cell differentiation takes place in the bone marrow, after which the progenitor cells enter the circulation and mature once reaching their target organ. Early results from single-cell culture experiments and colony-forming assays have produced the classic hierarchical tree model of hematopoiesis. The introduction of high-throughput, single-cell RNA sequencing is now revolutionizing our understanding of the differentiation process, questioning the classic tree-based models. By integrating the results from early cell culture experiments with single-cell transcriptomics, we present a differentiation landscape model of hematopoiesis and discuss it with focus on mast cells. The review also describes how the hematologic neoplasm systemic mastocytosis can be used to model human hematopoiesis using naturally occurring cell barcoding by means of the common KIT D816V mutation.
Introduction
Mast cells are tissue-resident cells, characterized by their high granularity and ability to release a large number of potent inflammatory mediators. The cells are well-known for their detrimental role in allergic diseases as well as in the hematologic neoplasm systemic mastocytosis.1 Even though mast cells develop from hematopoietic progenitors, they are often omitted or forgotten in models of hematopoiesis.
Hematopoiesis is classically represented using a treelike structure, with the hematopoietic stem cells as the tree stem, intermediate multi- and unipotent progenitors as branches, and mature cells as the leaves (Figure 1A). Single-cell transcriptomics is currently revolutionizing the understanding of blood cell formation, and the results suggest that the hematopoietic system is better visualized using a differentiation landscape. In this review, we describe mouse and human hematopoiesis with focus on the mast cell differentiation and maturation. We also discuss how naturally occurring cell barcoding in systemic mastocytosis can be used to resolve the human mast cell differentiation in vivo.
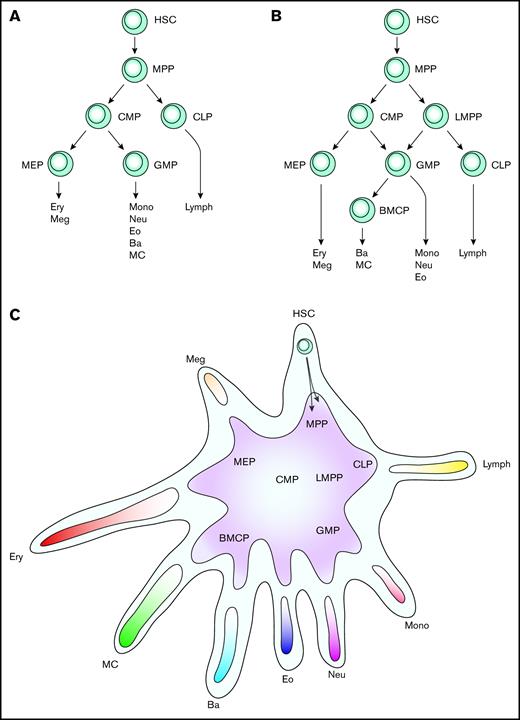
Evolution of models describing hematopoiesis. (A) Classic hierarchical model of hematopoiesis. (B) Revised model of hematopoiesis with LMPPs and BMCPs. (C) Differentiation landscape model of hematopoiesis. Hematopoietic stem and progenitor cells differentiate along ≥1 trajectories in the landscape. Each arm represents the entry point to 1 lineage, with the exception of the HSC arm, which is the starting point. Previously described bi- and multipotent progenitors are indicated, but the model is flexible and allows for tripotent erythroid, mast cell, and basophil progenitors, for example. This model focuses on myeloerythroid cell differentiation; the multiple lymphoid populations that exist are therefore not described in detail. Ba, basophil; CLP, common lymphoid progenitor; Eo, eosinophil; Ery, erythrocyte; HSC, hematopoietic stem cell; lymph, lymphocyte; MC, mast cell; Meg, megakaryocyte; mono, monocytes; MPP, multipotent progenitor; Neu, neutrophil.
Evolution of models describing hematopoiesis. (A) Classic hierarchical model of hematopoiesis. (B) Revised model of hematopoiesis with LMPPs and BMCPs. (C) Differentiation landscape model of hematopoiesis. Hematopoietic stem and progenitor cells differentiate along ≥1 trajectories in the landscape. Each arm represents the entry point to 1 lineage, with the exception of the HSC arm, which is the starting point. Previously described bi- and multipotent progenitors are indicated, but the model is flexible and allows for tripotent erythroid, mast cell, and basophil progenitors, for example. This model focuses on myeloerythroid cell differentiation; the multiple lymphoid populations that exist are therefore not described in detail. Ba, basophil; CLP, common lymphoid progenitor; Eo, eosinophil; Ery, erythrocyte; HSC, hematopoietic stem cell; lymph, lymphocyte; MC, mast cell; Meg, megakaryocyte; mono, monocytes; MPP, multipotent progenitor; Neu, neutrophil.
Are murine mast cells derived from bone marrow hematopoietic stem cells?
Blood cells are formed in distinct waves during embryogenesis. The yolk sac produces primitive erythroid progenitors followed by early erythromyeloid progenitors, whereas the aorta-gonad-mesonephros (AGM) is the first source of hematopoietic stem cells, which subsequently colonize the fetal liver and the bone marrow. Progenitors with mast cell–forming capacity are generated in the yolk sac.2,3 Recent fate-mapping experiments suggest that these progenitors give rise to the first skin mast cells in the embryo.4 These skin mast cells are then replaced by AGM-derived mast cells postnatally.4 Whether the 2-waved differentiation is specific for connective tissue-type and not mucosal mast cells is yet to be determined.
A seminal study from 1977 demonstrated that mouse bone marrow reconstitutes numerous mast cell compartments of irradiated mice, suggesting a bone marrow origin of mast cells in adult mice.5 However, the difficulties with reconstituting some mast cell compartments have questioned the role of bone marrow for the production of mast cells. Mast cell depletion followed by shielded irradiation and bone marrow reconstitution revealed that bone marrow progenitors contribute to only a small fraction of the peritoneal mast cell pool and do not give rise to skin or tongue mast cells in the shielded regions.4 Alternative progenitor sources could include white adipose tissue, from which cells can give rise to mast cells in tissues such as the intestine, skin, and white adipose tissue.6 Local mast cell proliferation and the pool of stem and progenitor cells in blood and peripheral tissues possibly also contribute to the mast cell numbers; however, the contribution of bone marrow progenitors for the formation of mast cells should not be dismissed. For example, Kanakura et al7 established a system in which irradiated and bone marrow reconstituted mice were depleted of peritoneal mast cells by distilled water injection. Indeed, virtually all mast cells that appeared in the peritoneum were of donor origin.7 The conclusion that bone marrow progenitors form peritoneal mast cells is also supported by observations in rats.8 Furthermore, several independent transfer experiments reveal that bone marrow progenitors give rise to mast cell progenitors in the lungs9-11 and small intestine.12,13
The seemingly contradictory results of the mast cell origin are likely because of the different experimental models used in the studies described previously. The bone marrow restores mast cells that have been depleted locally and contributes to the increase in tissue mast cell numbers on provocation. In contrast, some organs are partially reconstituted, whereas others obtain abnormally high numbers when transplanted bone marrow cells are transferred to mice with systemic mast cell deficiency.14 These observations could reflect a deregulation in the mast cell progenitor migration and maturation. Furthermore, peripheral mast cells are long-lived radioresistant cells that inhibit the recruitment and maturation of the mast cell progenitors from bone marrow.7 This likely explains the low reconstitution in some organs after transplantation of bone marrow cells into irradiated wild-type mice.5
Altogether, there is compelling evidence that bone marrow progenitors contribute to mast cell numbers in peripheral tissues of adult mice, especially upon tissue provocation, although local production of mast cells is likely important under steady-state conditions.
Identification of committed mast cell progenitors in mice
Unipotent (committed) mast cell progenitors were first identified in fetal blood in the 1990s.15 Several studies followed that described and validated the existence of committed mast cell progenitors in adult bone marrow,16,17 peripheral blood,18 intestine,19 and peritoneal cavity.20 Integrin α4β7 mediates migration of mast cell progenitors to peripheral tissues.12,21 Thus, high expression of integrin β7 subunit is commonly used to enrich for progenitors with mast cell–forming capacity. The integrin β7 expression is transient and is lost on maturation,11,20,22,23 making it suitable and specific for progenitor identification. Mast cell progenitors also express other surface markers including CD16/32 and c-Kit, which are commonly used to enrich for these cells.16,18,20 Notably, mast cell progenitors have a more mature phenotype in BALB/c mice than C57BL/6 mice, as assessed by FcεRI expression.18 Results obtained from different mouse strains may therefore not be immediately comparable, and flow cytometry gating strategies should be verified when using new mouse strains.
Identification of early progenitors with mast cell–forming capacity in mice
The differentiation trajectory from hematopoietic stem cells to mast cells is one of the long-withstanding questions in the mast cell field. The traditional strategy to resolve the differentiation trajectories of all cell types is to analyze single progenitors for multilineage output and determine which combinations of mature cells are possible. Experiments from the 1980s demonstrate that colonies of colony-forming unit–granulocyte, erythroid, macrophage, megakaryocyte contain mast cells, showing that mast cells derive from hematopoietic progenitors.24,25 Colony assays of spleen cells further reveal a close relationship between the mast cell and monocyte trajectories.26,27 Mixed neutrophil-monocyte-mast cell colonies are found, whereas mixed mast cell–erythroid colonies are only observed together with granulocytes and/or monocytes.26,27 These observations suggest that mast cell differentiation is closely coupled with granulocyte and monocyte development. A groundbreaking study by Arinobu et al later demonstrated that Lin− c-Kit+ integrin β7hi CD16/32hi cells in the spleen constitute bipotent basophil/mast cell progenitors (BMCPs),19 and the existence of splenic BMCPs has been verified by several studies.28-30 In addition, we recently identified Lin− c-Kit+ Sca-1− integrin β7hi CD16/32hi progenitors in mouse bone marrow.31 These bone marrow progenitors produce mast cells, basophils, and mixed mast cell–basophil colonies and are likely the precursors of the splenic BMCP population. One study did not reproducibly observe basophil potential in the splenic BMCP cell fraction, but instead reports mast cell potential only.32 This discrepancy is still to be resolved.
Further research to reconstruct the mast cell differentiation trajectory identified progenitors with dual mast cell–eosinophil output from Gata1 transgenic reporter mice. This cell output is specifically found in the Gata1-EGFP+ Lin− Sca-1− c-Kit+ CD41− CD16/32− CD105− CD150− (termed GE+ pre-GM) cell fraction.33 Basophil formation was not analyzed in the study, making it difficult to resolve a hierarchical differentiation. However, it is likely that GE+ pre-GM progenitors are less mature than the BMCPs, exemplified by the absence of CD16/32 in the GE+ pre-GM. A description of the transcription factors that regulate cell fate decisions is outside the scope of this review and is reviewed elsewhere.34,35
Single-cell transcriptomics reveals the mast cell differentiation trajectory in mice
Transcriptome analyses of bulk-sorted primary mature cells reveal that mast cells have a unique phenotype compared with cell types such as basophils, eosinophils, and neutrophils.36 Insights into the mast cell maturation can also be gained by analyzing the gene expression of bulk-sorted primary mast cell progenitors and mature mast cells.20,37 However, the deciphering of mast cell differentiation requires single-cell resolution. Recent advances in single-cell RNA sequencing are revolutionizing the research in the hematopoiesis field. Single-cell gene expression analysis reveals insights into the differentiation of most myeloerythroid cell types38-40; however, the mast cell differentiation trajectory has been notoriously difficult to resolve. Gene expression analysis of 4763 c-Kit+ hematopoietic progenitors is still not sufficient to distinguish the basophil and mast cell trajectories.41 We therefore performed RNA sequencing of >44 000 Lin− c-Kit+ and Lin− c-Kit+ Sca-1+ hematopoietic stem and progenitor cells.31 This resolves the differentiation from hematopoietic stem cells to the mast cell, basophil, eosinophil, neutrophil, monocyte, lymphoid, megakaryocyte, and erythroid lineage trajectories. Force-directed k-nearest neighbor visualization reveals that the mast cell and basophil entry points are positioned next to each other. Single-cell cultures and deep RNA sequencing confirm the existence of a common bipotent basophil/mast cell progenitor and position the cells at the basophil and mast cell entry points. Notably, a small fraction of the more mature single bone marrow BMCPs coexpress the Cma1 and Mcpt8 genes, which often are considered to be mast cell– and basophil-specific, respectively.31 These results suggest that a subset of the BMCPs is primed toward both cell lineages. They also indicate that Cma1 and Mcpt8 are not uniquely expressed in each respective lineage, consistent with observations of the IC-2 mouse cell line.42
W41/W41 mice have a point mutation in the Kit gene, which causes impaired c-Kit signaling. Single-cell RNA sequencing of Lin− c-Kit+ progenitors reveals that these mice do not have a specific mast cell differentiation trajectory, consistent with mast cell deficiency.31 W41/W41 mice still have BMCPs, confirmed by both single-cell transcriptomics and cell culture, pinpointing the differentiation block to the stage at which the progenitors commit to become mast cell.31
Is the mast cell differentiation trajectory associated with erythroid or granulocytic differentiation in mice?
Whether mast cells develop along the erythroid or granulocyte lineage trajectories is still debated. Cells sorted from the Lin− Sca-1+ c-Kit+ Ly6c− FcεRI− Thy1.1− Flk2+ multipotent progenitor gate form mast cells, some without a single-cell division.16 Chen et al16 therefore suggest that mast cells differentiate immediately from multipotent progenitors. Another study shows that the Lin− c-Kit+ Sca1lo CD27+ Flk-2− common myeloid progenitor (CMP) fraction has mast cell potential, whereas the Lin− c-Kit+ Sca1lo CD27+ Flk-2+ CD150−/lo granulocyte/monocyte progenitor (GMP) fraction does not.43 This can be interpreted as if mast cell progenitors differentiate separately from the GMPs and instead branch off at the CMP stage. The same study goes further and suggests that the mast cell differentiation follows the megakaryocyte and erythroid trajectories. This hypothesis is based on the similar gene expression patterns of single mast cell and erythroid-megakaryocyte progenitors, analyzing Mpo, Gfi1, Cebpa, Cpa3, Spi1, Gata2, Il1rl1, Gata1, and Epor.43 Our single-cell gene expression data, in which >2500 genes are detected per cell, places the mast cell entry points between the erythroid and the basophil lineages.31 Notably, cultures of bone marrow BMCPs do not produce erythroid cells.31 However, the erythroid trajectory was recently linked to the mast cell and/or basophil differentiation. Specifically, single-cell cultures of c-Kit+ CD55+ CD49fint CD105− CD41− CD150− progenitors produce colonies of erythroid cells and FcεRI+ cells.41 The identity of these FcεRI+ cells is yet to be determined, but they likely constitute basophils and/or mast cells. In contrast, a small fraction of single GMPs give rise to mixed neutrophil, mast cell, and basophil colonies19 and to mast cell, basophil, neutrophil, monocyte colonies.30 This appears conflicting at first glance, but may be a limitation in the model of hematopoiesis that is commonly used, a tree structure model (Figure 1A-B). We therefore propose that the mast cell differentiation and hematopoiesis in general is to be visualized using a hematopoietic landscape model (Figure 1C). In this model, a given cell in or adjacent to the hematopoietic stem cell compartment has potential to form all lineages. Moving along a differentiation trajectory increases the probability of a progenitor to form a specific cell type, rising to 100% when passing a lineage's entry point. A progenitor's potential to form multiple cell types is determined by studying the adjacent cell lineage trajectories. The hematopoietic landscape model is still compatible with the view of dividing cells into megakaryocyte/erythroid progenitors (MEPs), GMPs, and lymphoid-primed multipotent progenitors (LMPPs) (Figure 1B-C). It also explains the strong association between the mast cell and basophil lineages, the existence of mast cell, basophil, and neutrophil tripotent progenitors, and erythroid progenitors with residual basophil and/or mast cell–forming capacity.
The branching of CMPs into MEPs and GMPs is elegant,44 but the classic definition of GMPs can cause problems in understanding cell differentiation. The classification of all granulocytes (basophils, eosinophils, neutrophils, and perhaps mast cells despite not being found in circulation) in a common group assumes that they share differentiation trajectories. However, separating BMCPs from the classic GMPs indicates that the “granulocyte” in GMP mostly constitutes neutrophils.31 Further experiments will show if neutrophil/monocyte progenitors would more accurately describe the progenitors in the classic GMP gate. Nonetheless, the hematopoietic landscape points toward a more flexible differentiation than the tree models and allows combinations such as neutrophil/basophil progenitors and neutrophil/monocyte progenitors to occur in parallel.
Identification of early progenitors with mast cell–forming capacity in human
A bone marrow transplantation case report provides evidence that human mast cells are generated from hematopoietic stem cells.45 Although mast cells were of recipient origin at 4 months posttransplantation, donor-derived mast cells were apparent after 7 months and onwards.45 In vitro studies present further insights into the mast cell origin. CD34+ hematopoietic progenitors from bone marrow give rise to mast cells.46 Early theories that mast cell differentiate from basophils and CD14+ monocytes have been disproven, and the mast cell–forming capacity of CD34− cells is negligible compared with CD34+ progenitor cells in cord blood.47 It has also been shown that mast cells derived from CD34+ progenitors in vitro have potential to express a wide range of proteases such as tryptases, chymase, carboxypeptidase A3, granzyme B, and cathepsin G.48 However, it remains to be investigated if single CD34+ progenitors can give rise to tryptase-positive as well as chymase and tryptase double-positive mast cells, 2 classically defined mast cell subsets, in parallel.
The expression of c-Kit, the interleukin-3 (IL-3) receptor (CD123), FcεRI, and CD203c has been studied in detail during mast cell differentiation, which has facilitated the identification, isolation, and culture of primary mast cell progenitors. Stem cell factor promotes hematopoietic progenitors from various sources to form mast cells in vitro.49-52 In agreement, c-Kit is found throughout the development.53-55 Mast cells are favored by long-term cultures with stem cell factor, conditions that poorly support other lineages; therefore, caution needs to be exercised to support mast cells and other lineages simultaneously when performing colony assays.
Stem cell factor is important for late mast cell differentiation. However, the idea that stem cell factor and c-Kit signaling is indispensable for early mast cell differentiation was recently challenged.56 IL-3 and IL-6 are sufficient to stimulate the mast cell progenitors to form granules and express tryptase without c-Kit signaling.56 The classic distinction between basophil and mast cells in humans based on the lineage-promoting effects of IL-3 and stem cell factor, respectively, is therefore now less clear. However, recent data suggest that the human system is similar to mouse, where IL-3 induces mast cell differentiation.57 Some results suggest that CD123 expression in human is continuously downregulated during mast cell differentiation of cord blood cultures.54 Other studies show that CD123 is maintained during development and that IL-3 itself can induce the expression of the receptor in mature mast cells.53,55 Absence of CD123 on mast cells from the lung, skin, tonsil, and uterus, but stimulatory effects of IL-3 on intestinal mast cells likely suggest that the local environment plays an important role for receptor expression.58-60 FcεRI is one of the hallmark receptors of mast cells. Nonetheless, expression of the receptor is detected only on a subset of the human mast cells in vivo.61 This observation could possibly reflect the IL-4 and immunoglobulin E (IgE) levels in the microenvironment because these factors induce FcεRI expression on mast cells derived from cord blood in vitro.62,63 CD203c is expressed on basophils and mast cells and their CD34+ progenitors.64 Cell culture experiments of CD34+ CD203c+ bone marrow progenitors show evidence of mast cell–forming potential and considerable basophil-forming potential; however, the mast cell–forming potential is comparable in the CD34+ CD203c− and CD34+ CD203c+ fractions.64 Still, recent data suggest that CD203c in combination with other markers enrich for mast cell progenitors,65 likely the more differentiated ones, although this needs to be verified experimentally.
The mast cell–forming capacity is enriched in the CD34+ c-Kit+ CD13+ cell fraction of bone marrow and blood.66 Recently, it was demonstrated that Lin− CD34+ c-Kitint/hi FcεRI+ progenitors in peripheral blood form tryptase-positive c-Kit+ FcεRI+ cells, consistent with a mast cell phenotype, when cultured in conditions that stimulate myeloerythroid differentiation.56,67 An independent study showed that these progenitors fully agree in phenotype and frequency with the CD34+ c-Kithi HLA-DR−/int CD203c+ gating strategy used for identifying a putative peripheral blood mast cell progenitor.65 The Lin− CD34+ c-Kitint/hi FcεRI+ cells constitute unipotent (or almost committed) progenitors, which are further characterized by the expression of CD123 and integrin β7 and absence of CD45RA and CRTH2, placing the cells in the classic CMP sorting gate.56,67 However, this does not necessarily mean that mast cells are derived from CMPs.
The CD34+ CD38+ cell fraction in cord blood is enriched for mast cell–committed progenitors compared with CD34+ CD38− cells.68 Similarly, a higher frequency of pure mast cell colonies are found in the CD34+ HLA-DR− fraction compared with CD34+ HLA-DR+ cells, altogether suggesting that the unipotent mast cell progenitors are CD34+ CD38+ HLA-DR− cells68; however, it is likely that this population constitutes mast cell progenitors only.
Peripheral blood progenitors producing colonies with basophil- and/or mast cell–like phenotype were identified in the early 1980s.69 This study was undertaken before the discovery of stem cell factor and the establishment of optimal mast cell–promoting conditions; it is therefore possible that many or all these cells would be classified as basophils today. However, the observations still raise the question if human mast cells and basophils share an early differentiation trajectory. More recent data from cord blood cultures show that different subpopulations of CD34+ progenitors have potential to form mixed colonies including cells of the mast cell lineage.68 Single-cell cultures of 1200 CD34+ CD38+ progenitors, in which the resulting colonies are split and separately cultured in mast cell– or granulocyte/monocyte-promoting conditions, revealed 1 instance of a bilineage mast cell and basophil output.68 Similarly, 1 progenitor had trilineage mast cell/basophil/eosinophil output.68 Even though we cannot draw conclusions from these 2 observations, especially because erythroid differentiation is not investigated, it might suggest that the human and mouse mast cell differentiation trajectories are similar.
In contrast to the idea of a shared basophil/eosinophil/mast cell differentiation trajectory in human, a subpopulation of CD34+ c-Kit+ CD13+ bone marrow and peripheral blood progenitors that gives rise to mixed mast cell–monocyte colonies has also been described.66 Similarities in gene expression of CD34+ progenitors and mast cells indicate an alternative scenario in which mast cells develop immediately from hematopoietic stem cells.70 However, observations that mast cells are found in mixed colonies and the identification of bipotent mast cell/monocyte progenitors suggest that this is not the only developmental pathway.47,66,68 To summarize, the available data are not sufficient to draw definite conclusions on the human mast cell differentiation trajectory, and more studies on the topic are awaited.
Single-cell transcriptomics in human
Gene expression data of primary bulk-sorted mast cell progenitors and mature mast cells are available.67,70 Even though the data are valuable in many ways, it does not give significant insights into the mast cell differentiation trajectory. Single-cell RNA sequencing of >1000 human hematopoietic stem and progenitor cells from bone marrow show differentiation trajectories to the megakaryocyte, erythroid, neutrophil, monocyte/dendritic cell, and B-cell lineages.71 Hierarchical clustering also reveals a small population of progenitors with a phenotype suggesting that the cells are precursors of basophils, eosinophils, and/or mast cells. Genes highly expressed in these cells include CLC, PRG2, and HDC. The cell-forming potential of these progenitors is yet to be verified.
Gene expression analysis of almost 20 000 single CD34+ human cord blood cells have produced a map of human hematopoiesis, similar to that generated from bone marrow.72 CD34+ c-Kit+ FcεRI+ cells from bone marrow were sorted and sequenced and projected onto the basophil, eosinophil, and/or mast cell cluster.72 Notably, only CD34+ c-Kitint/hi FcεRI+ cells from peripheral blood have been confirmed to represent mast cell progenitors by in vitro culture.67 Unlike many other cell types, mast cells mature once reaching the peripheral tissues, and their progenitors are therefore found in the circulation. Thus, CD34+ c-Kit+ FcεRI+ cells in bone marrow do not necessarily constitute mast cell progenitors. In bone marrow, there are likely several cell types that can express the combination of CD34, c-Kit, and FcεRI, including basophils. The cell-forming potential of the CD34+ c-Kit+ FcεRI+ bone marrow progenitors should therefore be verified before concluding that these cells are mast cell progenitors.
Although single-cell RNA sequencing experiments suggest the structure of the differentiation landscape, the results need to be verified experimentally by in vitro colony-forming assays and/or in vivo lineage tracing experiments. The lack of understanding human mast cell progenitor differentiation probably is due to their poor proliferation potential. One sorted mast cell progenitor in culture for 1 week yields 3 cells in total (median).67 Non-mast cell lineage cells therefore easily outcompete mast cells in colony-forming assays and single-cell cultures. Thus, there is a great need for other means of analyzing progenitors with multilineage potential for mast cell output.
Naturally occurring cell barcoding in systemic mastocytosis may resolve the human mast cell differentiation
Systemic mastocytosis is a hematologic neoplasm characterized by mast cell accumulation in extracutaneous tissues. Patients with advanced disease may even display circulating mast cells, which are virtually undetectable in healthy subjects.73,74 The mast cell phenotype in systemic mastocytosis is commonly aberrant, which includes atypical morphology and expression of CD2 and CD25.74,75 Most patients have elevated serum tryptase levels and the spontaneously appearing c.2447A>T (D816V) mutation in the KIT gene. The mutation is commonly found in mast cells,65 but can in some patients also be detected in other lineages including lymphocytes, monocytes, neutrophils, basophils, and eosinophils.65,76-79 The spontaneous KIT mutation likely arises once in a given patient and then spreads by cell proliferation. For the mutation to appear in several lineages in the same individual likely requires the original mutation to arise in CD34+ multipotent progenitors. Assuming that these premises are correct, one can model human hematopoiesis by studying the distribution of the KIT mutation across cell types. This is analogous to barcoding experiments in which stem and progenitor cells are retrovirally transduced and lineage tracing is performed to decipher the differentiation trajectories.
In 1 study, patients with the KIT mutation in basophils, neutrophils, and monocytes were identified.77 More strikingly, 2 patients were found in which the mutation was detected in neutrophils or monocytes, but not basophils.77 These results may suggest that the human mast cell trajectory is more closely related to that of neutrophils and monocytes. The presence of the KIT mutation in monocytes but absence in neutrophils and vice versa, however, makes this interpretation difficult.65,77 One possibility is that the mutation arises in progenitors close to the mast cell entry point in the hematopoietic landscape in some patients, and in multipotent progenitors in others. Another simple explanation to the phenomenon is that not all cells within a cell lineage carry the KIT mutation. This can be exemplified by the results of a study by Mayado et al,65 in which an allele-specific oligonucleotide quantitative polymerase chain reaction, with a sensitivity of 0.01%, was used to detect the KIT mutation in several mature cell types. The results show that the frequency of the KIT mutation can differ an order of magnitude between 2 cell types that both are mutated within the same patient.65 The varying frequencies of affected cells could be a result of for example the time from disease onset and the proliferation rate of the malignant clones in relation to normal cells. Notably, it is yet to be determined whether the KIT mutation in some patients arises in unipotent mast cell progenitors or mast cells and in other patients in CD34+ hematopoietic stem/progenitor cells. For example, there are patients in which the KIT mutation is detected in mast cells only, whereas in others it can be detected in several cell lineages.65 It is possible that presence of the KIT mutation in multiple lineages is associated with a more severe disease phenotype.79
Because of the enucleation process during red blood cell maturation, the KIT mutation cannot be analyzed in mature erythrocytes; however, it has been identified in erythroid colonies in a case report.80 Studies of granulocyte/monocyte colonies have also been performed, and the KIT mutation can be detected in these cells in some patients as well.80,81 These results are compatible with reports of mutated CD34+ progenitors.65,79
The KIT mutation was recently described in a population of putative mesenchymal stem cells in systemic mastocytosis, suggesting that the mutation event might happen upstream the formation of the mesenchymal and hematopoietic stem cells in some patients.82 Another possible explanation is that the mutated cells gain potential to trans-differentiate into another cell type.83 Future studies will hopefully clarify these important findings.
Somatic mutations in genes such as JAK2, SRSF2, and TET2 are also found in some patients with systemic mastocytosis.81,84 These non-KIT mutations could therefore also be used as barcodes to study hematopoietic differentiation. The D816V point mutation in the KIT gene is well characterized and present in most patients, making it favorable compared with, for example mutations in TET2, where the mutation site differs between patients.85
To conclude, naturally occurring “barcodes” in the form of somatic mutations have the potential to reveal important insights into human hematopoietic differentiation.
Conclusion
The insight that mouse mast cells arise in 2 distinct waves in the embryo may change our understanding of mast cell differentiation in humans, especially in the context of mastocytosis. Aberrancies in the yolk sac–derived mast cells that decrease postnatally could then explain the favorable prognosis of cutaneous mastocytosis compared with the systemic disease. The cutaneous form is frequently observed in pediatric mastocytosis that often resolves before puberty,86 a phenomenon that occurs in parallel with the postnatal decline in yolk sac–derived mast cells. AGM-derived mast cells increase in numbers over time, and aberrancies in these cells may cause systemic disease.
It is clear the mast cells have potential to differentiate from bone marrow progenitors, and we propose that the classic tree model of hematopoiesis should be revised to a differentiation landscape to ensure compatibility between cell culture experiments as well as single-cell transcriptomics data. Although a clear understanding of murine mast cell differentiation is rapidly emerging, much work remains to be done in the human system, which is hampered by the poor in vitro proliferation potential of mast cell progenitors. We propose that investigating the cellular mutation distribution, a form of natural barcoding, may constitute a promising approach to decipher human in vivo mast cell differentiation.
Acknowledgments
This work was supported by the Swedish Research Council, the Swedish Cancer Society, Tore Nilson's Foundation for Medical Research, and Magnus Bergvall's Foundation.
Authorship
Contribution: J.G., J.S.U., G.N., and J.S.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Joakim S. Dahlin, Department of Medicine, Karolinska Institutet, SE-171 76 Stockholm, Sweden email: joakim.dahlin@ki.se.