Key Points
TLR activation suppresses expression of Fe-S cluster biogenesis factors NFS1, ISCU, HSC20, FXN, ISD11, GLRX5, CIAO1, FAM96A, and FAM96B.
Restriction of Fe-S cluster biogenesis not only impairs oxidative metabolism but also modulates histone and tubulin acetylation profiles.
Abstract
Given the essential roles of iron-sulfur (Fe-S) cofactors in mediating electron transfer in the mitochondrial respiratory chain and supporting heme biosynthesis, mitochondrial dysfunction is a common feature in a growing list of human Fe-S cluster biogenesis disorders, including Friedreich ataxia and GLRX5-related sideroblastic anemia. Here, our studies showed that restriction of Fe-S cluster biogenesis not only compromised mitochondrial oxidative metabolism but also resulted in decreased overall histone acetylation and increased H3K9me3 levels in the nucleus and increased acetylation of α-tubulin in the cytosol by decreasing the lipoylation of the pyruvate dehydrogenase complex, decreasing levels of succinate dehydrogenase and the histone acetyltransferase ELP3, and increasing levels of the tubulin acetyltransferase MEC17. Previous studies have shown that the metabolic shift in Toll-like receptor (TLR)–activated myeloid cells involves rapid activation of glycolysis and subsequent mitochondrial respiratory failure due to nitric oxide (NO)–mediated damage to Fe-S proteins. Our studies indicated that TLR activation also actively suppresses many components of the Fe-S cluster biogenesis machinery, which exacerbates NO-mediated damage to Fe-S proteins by interfering with cluster recovery. These results reveal new regulatory pathways and novel roles of the Fe-S cluster biogenesis machinery in modifying the epigenome and acetylome and provide new insights into the etiology of Fe-S cluster biogenesis disorders.
Introduction
Iron-sulfur (Fe-S) clusters are cofactors essential for mediating electron transfer in the mitochondrial respiratory chain, enzymatic catalysis in aconitase ACO2, and lipoylation of the pyruvate dehydrogenase complex (PDHc) and the α-ketoglutarate dehydrogenase complex (αKGDHc).1,2 Thus, controlled Fe-S cluster biogenesis is important for the production of not only ATP but also metabolites such as acetyl-coenzyme A (acetyl-CoA), α-ketoglutarate, and NAD+, which are needed for cellular processes such as fatty acid synthesis and posttranslational modifications. Fe-S cluster biogenesis is a complex process involving dozens of proteins in eukaryotes.3,4 The early steps of cluster assembly involve a complex composed of the cysteine desulfurase NFS1, LYRM4/ISD11, and the acyl carrier protein ACP,5 which associates with the scaffold protein ISCU and the allosteric factor frataxin (FXN) to synthesize nascent Fe-S clusters that are transferred to target proteins or intermediate carriers with the help of the dedicated HSC20-HSPA9 chaperone complex and other accessory proteins, such as GLRX53 (supplemental Figure 1A; supplemental Table 1). Although decreased oxidative phosphorylation (OXPHOS) due to disruption of Fe-S proteins in the respiratory complexes and tricarboxylic acid cycle is the most common feature of Fe-S cluster biogenesis disorders,6 the growing list of tissue-specific manifestations suggests that additional targets and etiologies might be involved.
In order to identify additional effectors and regulatory pathways, we examined the repression of Fe-S cluster biogenesis factors in both myeloid and nonmyeloid cells. Studies in the last decade have shown that profound changes in cellular metabolism are essential to meet the energetic and anabolic demands of myeloid cells such as macrophages and dendritic cells (DCs) in response to stimulation with Toll-like receptor (TLR) agonists.7 Live-cell metabolic assays and metabolite profiles have shown that glycolysis is rapidly activated and mitochondrial respiration collapsed upon stimulation of macrophages and DCs with TLR agonists.8-13 The glycolytic shift and the increase in pentose phosphate pathway activity are important for providing ribose for nucleic acid synthesis and NADPH for fatty acid synthesis, reactive oxygen species (ROS) and NO production, and reduction of glutathione.8-10 Interestingly, studies in the murine myeloid cell line RAW264.7, murine bone marrow–derived macrophages (BMDMs), and human monocyte-derived macrophages indicated that NFS1 and ISCU levels are decreased by the TLR4 agonist lipopolysaccharide (LPS) and the cytokine interferon-γ (IFN-γ),14,15 although it was unexpected that myeloid cells would reduce the expression of these key Fe-S cluster biogenesis factors when the generation of nitric oxide (NO) was already causing damage to Fe-S clusters.16,17 Here, we demonstrate that reduced expression of Fe-S biogenesis factors results not only in a decrease in mitochondrial metabolic activities but also in changes in histone acetylation and methylation in the nucleus and tubulin acetylation in the cytosol.
Methods
Cell culture, macrophage culture, and immunoblotting
Cells were cultivated in Dulbecco’s modified Eagle medium with 10% fetal bovine serum, 2 mM glutamine, and antibiotic-antimycotic (Invitrogen) at 37°C in a humidified incubator containing 5% CO2. Mouse peritoneal macrophages were isolated and cultured in Dulbecco’s modified Eagle medium/F12 supplemented with 10% fetal bovine serum, 10 mM glutamine, and antibiotics as previously described.18 Μacrophages were stimulated with LPS (100-200 ng/mL) or poly(I:C) (50 μg/mL) in the presence or absence of IFN-γ (25-50 ng/mL). HeLa or A549 cells were transfected every 3 days using On-TARGET Plus siRNA SMART Pools and Dharmafect 1 from Dharmacon. Plasmid transfections of RAW264.7 cell suspension (1e6 cells/35-mm dish) were performed with lipofectamine 2000 (Invitrogen), and LPS/IFN-γ or poly(I:C)/IFN-γ were added 4 hours after transfection. Clustered regularly interspaced short palindromic repeats (CRISPR) knockout of HSC20 was performed as previously described.19 Cytoplasmic extracts were prepared using 1% Triton lysis buffer containing 2 mM citrate. Whole-cell lysates were prepared using RIPA buffer containing 2 mM citrate. For immunoblots of histones, histone extracts were prepared using reagents from Abcam (ab113476). All animal experiments had ethical approval from the National Institutes of Health Ethics Committee.
Activity assays and metabolite measurements
Pyruvate dehydrogenase (PDH) enzyme activity and lactate assay kits were obtained from Mitoscience. In-gel respiratory complex I (RCI) and RCII activity were performed as previously described.20 Mitochondrial and cytosolic aconitase activity were assessed by in-gel aconitase assays as described previously using nondenaturing gels composed of a separating gel containing 5% (mouse) or 7% (human) acrylamide, 132 mM Tris base, 66 mM borate, 3.6 mM citrate, a stacking gel containing 5% acrylamide, 67 mM Tris base, 33 mM borate, and 3.6 mM citrate, and a running buffer at 4°C containing 25 mM Tris, 96 mM glycine, and 3.6 mM citrate.21
Results
The TLR4 agonist LPS represses expression of many components of the Fe-S cluster biogenesis machinery
Fe-S cluster biogenesis factors have crucial roles in maintaining the functions of many proteins that are important for cellular metabolism, including mitochondrial aconitase (ACO2), the SDHB subunit of RCII, the NDUFS1 subunit of RCI, and ferrochelatase (FECH) in the heme biosynthetic pathway. Small interfering RNAs (siRNAs) that target NFS1 depleted not only NFS1 but also its binding partner, ISCU, which led to the decrease in protein levels of SDHB, NDUFS1, and FECH (Figure 1A). In comparison, transcript levels of these genes did not change much after NFS1 silencing (supplemental Figure 2), consistent with previous studies that showed that some Fe-S proteins, including SDHB, FECH, and biotin synthase, become more susceptible to degradation when Fe-S cluster biogenesis is impaired.22-24 Likewise, siRNAs that target ISCU decreased the levels of NFS1, SDHB, NDUFS1, and FECH proteins (Figure 1A). Although PDHc and αKGDHc do not contain Fe-S clusters, covalently linked lipoyl cofactors are required for their enzymatic function. Fe-S clusters are critical for the function of lipoyl synthase,25 and depletion of lipoyl synthase (supplemental Figure 1B), NFS1, or ISCU (Figure 1A; supplemental Figure 1B) resulted in decreased lipoylation of the PDH E2 subunit and decreased PDHc activity (supplemental Figure 1C). Depletion of NFS1 and ISCU also diminished the enzymatic activity of cytosolic (ACO1) and mitochondrial (ACO2) aconitases (supplemental Figure 1D). Ablation of other key Fe-S cluster biogenesis factors, such as HSC2019 or FXN, also led to a decrease in the expression of downstream targets, including SDHB and NDUFS1, and lipoylation of PDH and αKGDH (Figure 1B-C).
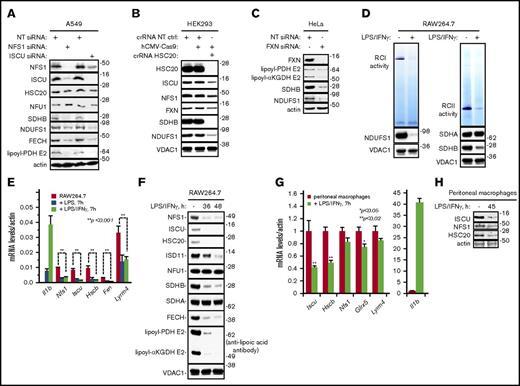
Reduced expression of Fe-S cluster biogenesis factors impairs the regeneration/repair of Fe-S proteins involved in mitochondrial oxidative metabolism. (A) Levels of lipoyl-PDH E2, SDHB, NDUFS1, and FECH in A549 cells decreased in response to silencing of NFS1 and ISCU. (B) Levels of SDHB and NDUFS1 decreased in CRISPR/Cas9 HSC20-knockout HEK293 cell mitochondrial lysates (crRNA HSC20). A scramble CRISPR RNA was used as control (crRNA NT CTRL). (C) Levels of lipoyl-PDH E2, lipoyl-αKGDH E2, SDHB, and NDUFS1 decreased in response to silencing of FXN. (D) Exposure of RAW264.7 cells to LPS and IFN-γ led to a decrease in RCI and RCII activity. (E) Activation of RAW264.7 cells with LPS or LPS/IFN-γ led to decreased mRNA levels of the Fe-S cluster biogenesis factors Nfs1, Iscu, Hscb, Fxn, and Lyrm4 (Isd11). (F) Protein levels of NFS1, ISCU, HSC20, ISD11, SDHB, and FECH and lipoylation of PDH E2 and αKGDH E2 decreased in RAW264.7 cells treated with LPS and IFN-γ. (G) Transcript levels of Iscu, Hsc20, Nfs1, Glrx5, and Lyrm4 decreased in peritoneal macrophages stimulated ex vivo with LPS and IFN-γ. (H) Protein levels of ISCU, NFS1, and HSC20 decreased in peritoneal macrophages stimulated ex vivo with LPS and IFN-γ. Actin or VDAC1 were used as loading controls. All data presented are representative of ≥3 independent experiments. NT, nontargeting.
Reduced expression of Fe-S cluster biogenesis factors impairs the regeneration/repair of Fe-S proteins involved in mitochondrial oxidative metabolism. (A) Levels of lipoyl-PDH E2, SDHB, NDUFS1, and FECH in A549 cells decreased in response to silencing of NFS1 and ISCU. (B) Levels of SDHB and NDUFS1 decreased in CRISPR/Cas9 HSC20-knockout HEK293 cell mitochondrial lysates (crRNA HSC20). A scramble CRISPR RNA was used as control (crRNA NT CTRL). (C) Levels of lipoyl-PDH E2, lipoyl-αKGDH E2, SDHB, and NDUFS1 decreased in response to silencing of FXN. (D) Exposure of RAW264.7 cells to LPS and IFN-γ led to a decrease in RCI and RCII activity. (E) Activation of RAW264.7 cells with LPS or LPS/IFN-γ led to decreased mRNA levels of the Fe-S cluster biogenesis factors Nfs1, Iscu, Hscb, Fxn, and Lyrm4 (Isd11). (F) Protein levels of NFS1, ISCU, HSC20, ISD11, SDHB, and FECH and lipoylation of PDH E2 and αKGDH E2 decreased in RAW264.7 cells treated with LPS and IFN-γ. (G) Transcript levels of Iscu, Hsc20, Nfs1, Glrx5, and Lyrm4 decreased in peritoneal macrophages stimulated ex vivo with LPS and IFN-γ. (H) Protein levels of ISCU, NFS1, and HSC20 decreased in peritoneal macrophages stimulated ex vivo with LPS and IFN-γ. Actin or VDAC1 were used as loading controls. All data presented are representative of ≥3 independent experiments. NT, nontargeting.
To further understand the physiological consequences of reduced Fe-S cluster biogenesis, we also examined the Fe-S cluster biogenesis machinery in myeloid cells that are activated with LPS and IFN-γ. In-gel activity assays showed that LPS/IFN-γ–activated RAW264.7 cells had significantly decreased RCI and RCII (Figure 1D) activity, consistent with previous studies that showed decreased mitochondrial oxygen consumption rates in LPS-activated myeloid cells.8,10 Previous studies have shown that the protein levels of NFS1 and ISCU decreased in RAW264.7 cells that were stimulated with LPS and IFN-γ.14 Therefore, we first examined the effect of LPS and IFN-γ on other components of the Fe-S cluster biogenesis machinery. Quantitative reverse transcription polymerase chain reaction analysis after 7-hour treatment of LPS and IFN-γ showed that messenger RNA (mRNA) levels of the core assembly factors Nfs1, Iscu, Hscb, Fxn, and Lyrm4 (Isd11) all decreased significantly (Figure 1E). Immunoblots also showed a marked decrease in the protein levels of not only NFS1, ISCU, HSC20, and ISD11 (Figure 1F) but also several accessory factors thought to be important for the transfer of nascent Fe-S clusters to recipient proteins, including GLRX5, CIAO1, FAM96A (also known as CIA2A), and FAM96B (or CIA2B) (supplemental Figure 1E), but not NFU1 (Figure 1F). Immunoblotting showed that LPS/IFN-γ–activated RAW264.7 cells had significantly decreased levels of SDHB, NDUFS1, FECH (Figure 1D,F), and ETFDH (supplemental Figure 1F), the Fe-S cluster containing oxidoreductase that links β-oxidation of fatty acids to OXPHOS.
To further confirm TLR-driven repression of Fe-S cluster biogenesis pathway, we isolated peritoneal macrophages and treated them with LPS and IFN-γ. Quantitative reverse transcription polymerase chain reaction showed that Iscu, Hscb, and Glrx5 transcript levels were significantly decreased after 7 hours of LPS/IFN-γ treatment (Figure 1G), and immunoblotting indicated that ISCU and HSC20 protein levels were decreased after 45 hours (Figure 1H). In addition, we stimulated RAW264.7 cells with poly(I:C), a TLR3 agonist that mimics viral infection, and levels of NFS1, HSC20, FAM96B, and GLRX5 were decreased, as were activity of RCI, RCII, and aconitases and levels of NDUFS1, SDHB, FECH, and lipoyl-PDH E2 (supplemental Figure 3A-E). Similarly, stimulation of RAW264.7 cells with IFN-γ and phorbol 12-myristate 13-acetate, a phorbol ester (PMA) that can activate NF-κB, led to decreased levels of NFS1, GLRX5, and FAM96B, along with decreased aconitase activity and lipoylation of PDH E2 and αKGDH E2 (supplemental Figure 3F). Thus, the Fe-S cluster biogenesis machinery was regulated in response to a variety of TLR agonists and chemical stimulants.
Restriction of Fe-S cluster biogenesis exacerbates NO-induced damage to Fe-S proteins in TLR-stimulated cells
Studies in DCs have shown that the rapid (<1 hour) increase in glycolytic metabolism upon stimulation with LPS was mediated by the AKT- and HIF-1α–dependent activation of glycolytic enzymes,10,26 whereas the prolonged commitment to glycolysis depended on the induction of inducible nitric oxide synthase (iNOS),17 which generates NO that inhibits mitochondrial electron transport and blocks mitochondrial ATP production. In RAW264.7 cells, the Akt/mTOR signaling pathway was activated within minutes of exposure to LPS and IFN-γ, as indicated by increased phosphorylation of Akt at both Thr308 and Ser473 and increased phosphorylation of ribosomal protein S6 at Ser 235/236, a downstream target of Akt/mTOR signaling (Figure 2A). Lactate production also increased rapidly, indicating that glycolysis increased in LPS/IFN-γ–stimulated cells (Figure 2B). In resting macrophages, NF-κB is sequestered in the cytoplasm through its association with inhibitors of κB (IκBs). Within 15 minutes of activation, IκBα was degraded, allowing the NF-κB dimer to translocate to the nucleus to increase the expression of iNOS, HIF-1α, and cyclooxygenase-2 (COX2) (Figure 2A). HIF-1α levels were elevated between 3 and 24 hours after exposure to LPS and IFN-γ, consistent with the activation of its transcriptional regulators mTOR and NF-κB and reduced prolyl hydroxylase–dependent degradation of HIF-α proteins. 9,27,28
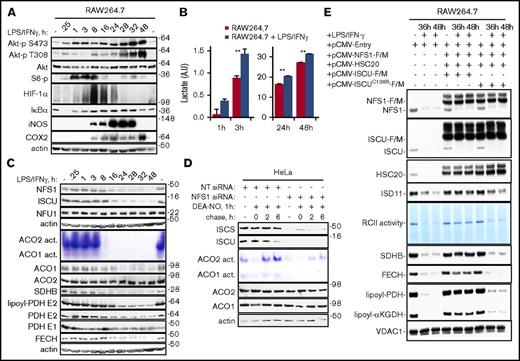
Repression of Fe-S cluster biogenesis factors amplifies NO-induced damage of Fe-S proteins. Exposure of RAW264.7 cells to LPS and IFN-γ led to rapid activations of the Akt, mTOR, HIF-1α, and NF-κB signaling pathways (A) and lactate production (B), followed by an induction of iNOS (A) and a concomitant decrease in the levels of NFS1, ISCU, SDHB, and FECH, aconitase activity, and lipoyl-PDH E2 (C). (D) HeLa cells transfected with NT siRNA or NFS1 siRNA were treated with 400 μM DEA-NO for 1 hour and then washed and incubated in fresh medium for up to 6 hours. In-gel assays indicated that aconitase activity recovered poorly in NFS1-depleted cells compared with control cells. (E) RCII activity and levels of SDHB, FECH, lipoyl-PDH, and lipoyl-KGDH were largely rescued by cotransfection of RAW264.7 cells with pCMV-NFS1-F/M, pCMV-ISCU-F/M, and pCMV-HSC20. In contrast, substitution of ISCU with a construct that expressed an inactive ISCU mutant (ISCU C138S) failed to restore RCII activity and the levels of SDHB, FECH, lipoyl-PDH, and lipoyl-KGDH. **P < .004.
Repression of Fe-S cluster biogenesis factors amplifies NO-induced damage of Fe-S proteins. Exposure of RAW264.7 cells to LPS and IFN-γ led to rapid activations of the Akt, mTOR, HIF-1α, and NF-κB signaling pathways (A) and lactate production (B), followed by an induction of iNOS (A) and a concomitant decrease in the levels of NFS1, ISCU, SDHB, and FECH, aconitase activity, and lipoyl-PDH E2 (C). (D) HeLa cells transfected with NT siRNA or NFS1 siRNA were treated with 400 μM DEA-NO for 1 hour and then washed and incubated in fresh medium for up to 6 hours. In-gel assays indicated that aconitase activity recovered poorly in NFS1-depleted cells compared with control cells. (E) RCII activity and levels of SDHB, FECH, lipoyl-PDH, and lipoyl-KGDH were largely rescued by cotransfection of RAW264.7 cells with pCMV-NFS1-F/M, pCMV-ISCU-F/M, and pCMV-HSC20. In contrast, substitution of ISCU with a construct that expressed an inactive ISCU mutant (ISCU C138S) failed to restore RCII activity and the levels of SDHB, FECH, lipoyl-PDH, and lipoyl-KGDH. **P < .004.
Interestingly, a marked decrease in NFS1 and ISCU levels coincided with a peak in iNOS induction, a decrease in aconitase activity and lipoylation of PDH E2, and a decrease in SDHB and FECH protein levels (Figure 2C, 8-48 hours). While the production of NO during macrophage activation has been shown to diminish the activity and levels of ACO2 and SDH,16 levels of functional Fe-S proteins also depend on the ability of cells to regenerate or repair damaged Fe-S proteins.21 In-gel activity assays indicated that aconitase activity decreased after exposure of HeLa cells to DEA-NO, an NO donor, consistent with NO-driven damage of the Fe-S clusters in ACO1 and ACO2 (Figure 2D). However, within 2 hours after the removal of DEA-NO, aconitase activity had recovered, indicating that Fe-S clusters were rapidly repaired/regenerated in normal HeLa cells. In contrast, the combination of NO and NFS1 siRNA resulted in even lower aconitase activity, indicating that restriction of Fe-S cluster biogenesis could exacerbate NO-mediated damage to Fe-S proteins (Figure 2D). Further corroborating this notion, overexpression of NFS1-FLAG/myc (NFS1-F/M), ISCU-F/M, and HSC20 in LPS/IFN-γ–treated RAW264.7 cells alleviated the loss of SDHB and FECH proteins and restored most of the RCII activity and PDHc and αKGDHc lipoylation (Figure 2E). Taken together, these results suggest that TLR-driven repression of Fe-S cluster biogenesis machinery can aggravate NO-mediated damage to Fe-S proteins by interfering with cluster recovery.
Oxidative stress, nitrative stress, and NF-κB signaling contribute to the repression of Fe-S cluster biogenesis, whereas inhibition of TLR-driven glycolysis alleviates the repression of NFS1 and ISCU
Next, we asked what might contribute to TLR-driven repression of Fe-S cluster biogenesis in RAW264.7 cells. Our previous studies showed that ISCU levels in fibroblasts and myoblasts rapidly decrease following H2O2 or high O2 treatment.29 Similarly, ISCU levels decreased significantly when RAW264.7 cells were cultured for 24 to 48 hours under high O2 conditions (Figure 3A) or treated with menadione for 1 hour (Figure 3B). These results, together with a marked decrease in ISCU and GLRX5 levels in cells exposed to the slow-releasing NO donor NOC-18 (Figure 3C; supplemental Figure 4A), suggested that, in addition to transcription repression, oxidative stress and iNOS induction may contribute to the restriction of Fe-S cluster biogenesis in activated macrophages. ISCU transcripts have previously been found to be the targets of miR-210, which is induced by HIF-1α.30 Therefore, we asked whether activation of HIF-1α (Figure 2A) decreased ISCU in LPS/IFN-γ–activated cells. When RAW264.7 cells were cultured at 0.5% O2, HIF-1α levels increased, but ISCU levels only slightly decreased (supplemental Figure 4B), suggesting that the previously reported HIF-1α /miR-210–mediated regulation of ISCU may not contribute much to the repression of ISCU in LPS/IFN-γ–stimulated RAW264.7 cells.
Oxidative and nitrative stress and NF-κB signaling contribute to repression of Fe-S cluster biogenesis, whereas inhibition of TLR-driven glycolysis reduces the repression of NFS1 and ISCU. (A-B) ISCU levels decreased when RAW264.7 cells were exposed to 95% oxygen (A) or menadione (B). (C) ISCU levels decreased when RAW264.7 cells were exposed to the NO donor (NOC-18). (D) Low glucose concentrations in the media mitigated the LPS/IFN-γ– or poly(I:C)/IFN-γ–induced suppression of NFS1 and ISCU and partially rescued lipoyl-PDH E2 and FECH levels and aconitase activity. (E) Repression of NFS1 was mitigated by pretreating RAW264.7 cells with MLN120B, an inhibitor of NF-κB signaling, for 1 hour before the addition of LPS/IFN-γ. (F) Levels of NFS1, SDHB, and NDUFS1 decreased in HeLa cells in response to TNF and IFN-γ.
Oxidative and nitrative stress and NF-κB signaling contribute to repression of Fe-S cluster biogenesis, whereas inhibition of TLR-driven glycolysis reduces the repression of NFS1 and ISCU. (A-B) ISCU levels decreased when RAW264.7 cells were exposed to 95% oxygen (A) or menadione (B). (C) ISCU levels decreased when RAW264.7 cells were exposed to the NO donor (NOC-18). (D) Low glucose concentrations in the media mitigated the LPS/IFN-γ– or poly(I:C)/IFN-γ–induced suppression of NFS1 and ISCU and partially rescued lipoyl-PDH E2 and FECH levels and aconitase activity. (E) Repression of NFS1 was mitigated by pretreating RAW264.7 cells with MLN120B, an inhibitor of NF-κB signaling, for 1 hour before the addition of LPS/IFN-γ. (F) Levels of NFS1, SDHB, and NDUFS1 decreased in HeLa cells in response to TNF and IFN-γ.
Previous studies have shown that inhibition of LPS-driven glycolysis with the hexokinase inhibitor 2-deoxyglucose (2-DG) suppresses LPS-induced interleukin-1β (IL-1β) and IL-10, but not tumor necrosis factor (TNF) production.9,12 We then asked whether limiting glycolysis might affect the LPS-induced repression of NFS1 and ISCU. Reducing glucose concentration in the media almost completely blocked the decline of NFS1 and ISCU levels in cells stimulated with LPS and IFN-γ or poly(I:C) and IFN-γ (Figure 3D, lanes 4, 5, 7, and 8). FECH protein, aconitase activity and lipoylation in PDH E2 were also partially rescued in media that contained lower levels of glucose (Figure 3D). Similar to glucose deprivation, 2-DG also alleviated the repression of NFS1 and ISCU and reduced the loss of PDH E2 lipoylation and aconitase activity (supplemental Figure 4C). Given that LPS, poly(I:C), and PMA are all known to modulate NF-κB signaling, we then tested whether NF-κB was involved in the regulation of Fe-S cluster biogenesis using MLN120B, which inhibits NF-κB activation via the inhibition of IKKβ.31 Indeed, MLN120B mitigated the decrease in NFS1 levels, suggesting that NF-κB might be involved in the regulation of NFS1 (Figure 3E). However, MLN120b treatment did not prevent LPS/IFN-γ–induced repression of ISCU, which may explain why MLN120b only partially prevented the decrease in FECH levels. NF-κB is a widely expressed transcription factor that is activated by a variety of stress and inflammatory stimuli, including TNF.32 In TNF/IFN-γ–treated HeLa cells, NFS1 levels decreased, as did PDH lipoylation and SDHB levels (Figure 3F), suggesting that restriction of Fe-S cluster biogenesis may contribute to the physiological cellular response to TNF.
Restriction of Fe-S cluster biogenesis impairs PDHc, ELP3, and SDH function, resulting in decreased overall histone acetylation levels and increased H3K9me3 levels
Acetyl-CoA is the substrate for acetyltransferase that catalyzes protein acetylation.33 Mitochondrial acetyl-CoA is generated by the decarboxylation of pyruvate by PDHc, β-oxidation of fatty acids, and the metabolism of branched-chain amino acids through the action of the branched-chain α-ketoacid dehydrogenase complex. As both PDHc and the branched-chain α-ketoacid dehydrogenase complex require lipoylation for their activity, we asked whether repression of Fe-S cluster biogenesis could decrease histone acetylation. Indeed, knockdown of NFS1 or PDH-E2 both resulted in an overall decrease in the acetylation of histones (Figure 4A-B), suggesting that restriction of Fe-S cluster biogenesis might mediate epigenetic changes by decreasing PDHc lipoylation. In addition, recent studies have shown that ELP3, an Fe-S cluster containing a subunit of the Elongator complex,34 is required for maintaining H3K9ac and H3K18ac levels.35,36 We observed a significant loss of ELP3 protein in NFS1-siRNA and ISCU-siRNA knockdown HeLa cells (Figure 4C-D), suggesting yet another mechanism for the decrease in histone acetylation observed in cells deficient in Fe-S cluster biogenesis factors. Interestingly, heterochromatinization is one of several mechanisms that reduces FXN expression in patients with the neurodegenerative disease Friedreich ataxia (FRDA).37 FXN-siRNA–treated cells also had reduced lipoyl-PDH E2, ELP3, and overall histone acetylation levels (Figure 4E), suggesting that repression of Fe-S cluster biogenesis may potentiate heterochromatinization in the FXN locus in FRDA patients.
Restriction of Fe-S cluster biogenesis impairs PDHc and ELP3 function, resulting in decreased overall histone acetylation. (A-B) Knockdown of NFS1 (A) or PDH E2 (B) resulted in decreased histone acetylation (ac-histones) in HeLa cells. (C-D) Knockdown of NFS1 (C) or ISCU (D) reduced levels of the acetyltransferase ELP3. (E) Knockdown of FXN decreased lipoylation of PDH E2 and αKGDH E2, decreased ELP3 levels, and decreased acetylation of histones. (F) Histone acetylation decreased in RAW264.7 cells activated with LPS/IFN-γ. (G) Acetate supplementation mitigated the LPS/IFN-γ–induced decrease in histone acetylation. (H) Treatment of HeLa cells with the iron chelator deferoxamine (Dfo) decreased levels of FXN, ISCU, NFS1, ISD11, lipoyl-PDH E2, ELP3, and SDHB, decreased overall histone acetylation, and increased H3K9me3 modifications.
Restriction of Fe-S cluster biogenesis impairs PDHc and ELP3 function, resulting in decreased overall histone acetylation. (A-B) Knockdown of NFS1 (A) or PDH E2 (B) resulted in decreased histone acetylation (ac-histones) in HeLa cells. (C-D) Knockdown of NFS1 (C) or ISCU (D) reduced levels of the acetyltransferase ELP3. (E) Knockdown of FXN decreased lipoylation of PDH E2 and αKGDH E2, decreased ELP3 levels, and decreased acetylation of histones. (F) Histone acetylation decreased in RAW264.7 cells activated with LPS/IFN-γ. (G) Acetate supplementation mitigated the LPS/IFN-γ–induced decrease in histone acetylation. (H) Treatment of HeLa cells with the iron chelator deferoxamine (Dfo) decreased levels of FXN, ISCU, NFS1, ISD11, lipoyl-PDH E2, ELP3, and SDHB, decreased overall histone acetylation, and increased H3K9me3 modifications.
Previous studies have shown that LPS regulates the expression of several proinflammatory genes, including Cox-2 and Cxcl2, by modulating histone deacetylase (HDAC) expression.38 In LPS/IFN-γ– and poly(I:C)/IFN-γ–activated RAW264.7cells, the overall histone acetylation levels decreased (Figure 4F; supplemental Figure 5A) at time points at which several Fe-S cluster biogenesis factors and lipoyl-PDH E2 also decreased significantly (Figure 2B), suggesting that decreased PDH activity due to NO and repression of Fe-S cluster biogenesis factors may also contribute to decreased histone acetylation in activated myeloid cells. Supporting this notion, supplementation with sodium acetate, which can be converted to acetyl-CoA by acyl-CoA synthetase short-chain family member 2,33 mitigated the LPS-induced decrease in histone acetylation (Figure 4G).
Methylation of lysine residues in histones represents another form of posttranslational modification that can either activate or repress transcription.39 Chromosome immunoprecipitation experiments have revealed changes in gene-specific H3 methylation patterns upon LPS stimulation.40 Interestingly, we observed an overall increase in the repressive histone mark H3K9me341 in the late phase of the LPS response (supplemental Figure 5B). Due to the structural similarity to α-ketoglutarate, succinate accumulation that results from loss of SDH activity can competitively inhibit α-ketoglutarate–dependent enzymes, including prolyl hydroxylases and Jumonji-C domain histone demethylases.42 Exposure of HeLa cells to dimethyl succinate or the SDH inhibitors 3-NPA and malonate increased H3K9me3 levels (supplemental Figure 5C). When cells were depleted of NFS1 using NFS1 siRNA, there was an increase in H3K9me3 levels, consistent with a loss of SDHB and inhibition of H3K9me3 demethylation (supplemental Figure 5D). These results suggested that repression of Fe-S cluster biogenesis might contribute to the deactivation of inflammatory genes by decreasing overall histone acetylation and increasing H3K9me3 modifications that promote heterochromatin formation and gene silencing.
Since our previous studies showed that iron deficiency not only limits the availability of iron for Fe-S cluster biogenesis but also causes a decrease in mRNA and protein levels of ISCU and FXN in RD4 and HEK293 cells,21,43 we asked whether iron deficiency also resulted in decreased histone acetylation and increased H3K9me3 levels. Indeed, in HeLa cells treated with the iron chelator deferoxamine, there was a decrease in overall histone acetylation and an increase in H3K9me3 levels along with decreased protein levels of several Fe-S cluster biogenesis factors, including FXN, ISCU, NFS1, and ISD11, decreased PDHc lipoylation, and decreased levels of ELP3 and SDHB (Figure 4H). Similarly, a TNF/IFN-γ–induced decrease in NFS1 and lipoyl PDH-E2 levels in HeLa cells correlated with a decrease in histone acetylation levels (supplemental Figure 5E). Thus, repression of Fe-S cluster biogenesis by a variety of stimuli, including TLR agonists, iron deprivation, and TNF/NF-κB signaling might effect epigenetic changes through the loss of PDHc, ELP3, and SDHB activity.
Repression of Fe-S cluster biogenesis induced acetylation of α-tubulin
In contrast to the decrease in histone acetylation, a recent study showed that microtubule acetylation increased following LPS treatment of RAW264.7 cells (Figure 5A), and knockdown of the cytosolic acetyltransferase MEC17 (also know as αTAT1) inhibited the LPS-induced tubulin acetylation and concomitantly decreased IL-10 production.44 Although the Elongator complex has been shown to be important for tubulin acetylation in cortical neurons, gene ablation studies indicated that MEC17 is the major tubulin acetyltransferase in most tissues in mice.45 Interestingly, we showed that silencing of FXN and ISCU resulted in increased MEC17 levels and increased α-tubulin acetylation (Figure 5B-C). Because acetyl-CoA is unstable and cannot readily cross organelle membranes, it has been postulated that distinct pools of acetyl-CoA are present in subcellular compartments, and localized production of acetyl-CoA by spatial control of acetyl-CoA producers (PDHc, ATP-citrate lyase, and acyl-CoA synthetase short-chain family member 2) confer spatial specificity to the metabolic regulation of acetylation.46 Thus, our data are consistent with a growing body of literature suggesting that spatial choreography of acetylation occurs in different subcellular compartments in response to cellular needs.
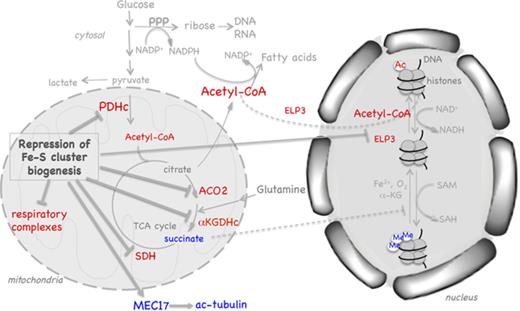
Restriction of Fe-S cluster biogenesis increases expression of the acetyltransferase MEC17 and induces hyperacetylation of tubulin. (A) LPS/IFN-γ activation of RAW264.7 cells increased acetylated α-tubulin (ac-tubulin) levels. (B-C) Levels of the cytosolic acetyltransferase MEC17 and tubulin acetylation increased in response to knockdown of FXN (B) or ISCU (C). (D) Schematic illustrating how restriction of the Fe-S cluster biogenesis machinery may increase (blue) or decrease (red) key metabolites and proteins important in metabolic and epigenetic remodeling.
Restriction of Fe-S cluster biogenesis increases expression of the acetyltransferase MEC17 and induces hyperacetylation of tubulin. (A) LPS/IFN-γ activation of RAW264.7 cells increased acetylated α-tubulin (ac-tubulin) levels. (B-C) Levels of the cytosolic acetyltransferase MEC17 and tubulin acetylation increased in response to knockdown of FXN (B) or ISCU (C). (D) Schematic illustrating how restriction of the Fe-S cluster biogenesis machinery may increase (blue) or decrease (red) key metabolites and proteins important in metabolic and epigenetic remodeling.
Discussion
Due to the many essential roles of Fe-S clusters in cellular function and the vulnerability of Fe-S proteins to oxidative and nitrative stress, the spectrum of diseases attributable to abnormal Fe-S cluster biogenesis/repair extends from FRDA, the most common form of hereditary ataxia, to GLRX5- and HSPA9-related sideroblastic anemias, ISCU myopathy, and several lethal multiple mitochondrial dysfunctions syndromes.6 Our study showed that in addition to disrupting OXPHOS, loss of Fe-S biogenesis factors also resulted in an overall decrease in histone acetylation, a hallmark of the transcriptionally silenced heterochromatized regions of DNA. In eukaryotes, key metabolites, including acetyl-CoA and succinate, are used not only as energy sources but also as substrates for posttranslational modifications and signaling molecules.33,39,47 Decreased levels of Fe-S cluster biogenesis factors led to the loss of SDH and PDHc activity, which can lead to a buildup of succinate and a decrease in acetyl-CoA levels.22,48 In addition to being an intermediate in the tricarboxylic acid cycle, succinate also acts as an inflammatory signal that induces IL-1β through HIF-1α activation in macrophages and as an oncometabolite in tumors driven by inactivating mutations in SDH.9,49 The PDHc-mediated conversion of pyruvate to acetyl-CoA represents a critical regulatory point in cellular energy metabolism, protein acetylation, and synthetic pathways for the production of fatty acids, cholesterol, and heme.33 The flux through PDHc has been shown to be tightly regulated by several well-documented mechanisms.50 Our study showed that control of Fe-S cluster biogenesis, whether caused by TLR or TNF signaling, iron deprivation, or oxidative/nitrative stress, may provide an additional layer of regulation of PDHc activity and acetyl-CoA metabolism.
Extensive chromatin immunoprecipitation data collected at the FRDA locus, which contains an expanded trinucleotide repeat (GAA)n in the first intron of FXN, had shown that the levels of the heterochromatin mark H3K9me3 were enriched, whereas the levels of acetylated H3 and H4 were reduced.37 Our finding that decreased Fe-S cluster biogenesis resulted in decreased overall histone acetylation and increased H3K9me3 levels poses an interesting possibility of a negative feedback mechanism that potentiates a progressive loss of FXN expression in the postmitotic cells that are most severely affected in FRDA. Furthermore, our results showed that silencing of Fe-S cluster biogenesis factors reduced the levels of ELP3, a subunit of the Elongator complex that has roles in growth cone motility and axonal outgrowth.34,35 In addition, our studies revealed that silencing of FXN or ISCU induced the α-tubulin acetyltransferase MEC17, resulting in hyperacetylation of α-tubulin. Reversible acetylation of tubulin confers mechanical protection to microtubules51 and controls their interaction with cellular components52 and is critical for neuronal development and function, growth factor or apoptotic signaling, and cell cycle progression.52 Acetylation of K40 of α-tubulin is mainly controlled by the cytosolic acetyltransferase MEC17 and the cytosolic deacetylases HDAC6 and SIRT2.52 Notably, a mouse model of FRDA cardiomyopathy with ablation of FXN had increased mitochondrial protein acetylation that was attributed to a decrease in the mitochondrial NAD+/NADH ratio, which can lower the activity of the mitochondrial deacetylase SIRT3.53 Thus, our findings suggest that the roles of PDHc, ELP3, and MEC17 in the etiology of FRDA, GLRX5-related sideroblastic anemias, and other Fe-S cluster biogenesis disorders warrant further study.
Our finding that silencing of Fe-S cluster biogenesis factors markedly decreases histone acetylation also suggest that repression of Fe-S cluster biogenesis might contribute to the complex epigenetic changes during myeloid cell activation54,55 (supplemental Figure 6). Epigenetic changes are important not only for the early hours of activation but also for the later stages when inflammatory genes are deactivated, as timely resolution is important to limit the detrimental effects of inflammation. Recent studies have suggested that histone deacetylation is an important mechanism in the deactivation of inflammatory genes.56,57 Furthermore, we showed that silencing of Fe-S cluster biogenesis could increase MEC17 levels and induce tubulin acetylation. Recent studies have shown that MEC17-mediated tubulin hyperacetylation in LPS-activated myeloid cells plays an important role in the induction of an anti-inflammatory cytokine, IL-10, which is critical for preventing tissue injury caused by excessive inflammation.44 Taken together, our findings suggest that repression of Fe-S cluster biogenesis could contribute to modulation of key components of the inflammatory response (supplemental Figure 6).
Despite the critical roles of Fe-S cofactors in many cellular functions, relatively little is known about the regulation of the Fe-S cluster biogenesis machinery. Interestingly, recent studies revealed that NFS1 expression was amplified in a subset of lung adenocarcinomas and positive selection of NFS1 may promote tumor formation by protecting tumor cells from undergoing ferroptosis in response to oxidative damage.58 Our results showed that suppression of the Fe-S cluster biogenesis machinery not only resulted in disruption of mitochondrial functions but also modulated the acetylation profile of histones and tubulin (Figure 5D), revealing previously unrecognized targets and signaling pathways that might be important for devising therapy for Fe-S cluster biogenesis disorders.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank their colleagues in the Rouault laboratory for constructive discussions.
This work was supported by the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development Intramural Research Program.
Authorship
Contribution: W.-H.T. designed the study, conducted experiments, and analyzed data; N.M., D.-L.Z., and E.M.P. performed experiments and analyzed data; H.O. and M.C.G. assisted with mouse experiments; and T.A.R. and D.W.M. supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tracey A. Rouault, National Institutes of Health, Building 35A, Room 2D-824, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: rouault@mail.nih.gov.