Key Points
Human platelets, endothelial cells, and plasma proteins are extensively O-glycosylated, with >1123 O-glycosites identified in this study.
O-glycosites can be classified into functional subgroups; one important function includes the protection from proteolytic processing.
Abstract
The hemostatic system comprises platelet aggregation, coagulation, and fibrinolysis, and is critical to the maintenance of vascular integrity. Multiple studies indicate that glycans play important roles in the hemostatic system; however, most investigations have focused on N-glycans because of the complexity of O-glycan analysis. Here we performed the first systematic analysis of native-O-glycosylation using lectin affinity chromatography coupled to liquid chromatography mass spectrometry (LC-MS)/MS to determine the precise location of O-glycans in human plasma, platelets, and endothelial cells, which coordinately regulate hemostasis. We identified the hitherto largest O-glycoproteome from native tissue with a total of 649 glycoproteins and 1123 nonambiguous O-glycosites, demonstrating that O-glycosylation is a ubiquitous modification of extracellular proteins. Investigation of the general properties of O-glycosylation established that it is a heterogeneous modification, frequently occurring at low density within disordered regions in a cell-dependent manner. Using an unbiased screen to identify associations between O-glycosites and protein annotations we found that O-glycans were over-represented close (± 15 amino acids) to tandem repeat regions, protease cleavage sites, within propeptides, and located on a select group of protein domains. The importance of O-glycosites in proximity to proteolytic cleavage sites was further supported by in vitro peptide assays demonstrating that proteolysis of key hemostatic proteins can be inhibited by the presence of O-glycans. Collectively, these data illustrate the global properties of native O-glycosylation and provide the requisite roadmap for future biomarker and structure-function studies.
Introduction
Mucin-type O-glycosylation, or N-Acetylgalactosamine (GalNAc) O-glycosylation is arguably the most prevalent and diverse form of O-glycosylation.1 GalNAc O-glycosylation (hereafter O-glycosylation) biosynthesis is initiated by a family of as many as 20 differentially expressed GalNAc-transferases (GalNAc-Ts)2 that add a GalNAc monosaccharide to selected serine (Ser), threonine (Thr), and, possibly, tyrosine (Tyr) residues.1,3 The major O-glycan structures in humans are the sialylated Core 1 O-glycans—sialyl T and disialyl-T—along with the lesser represented Core 2 glycans (Figure 1).4,5 These can be further elongated and modified, generating a large set of O-glycan structures.4-6 Historically, O-glycosylation has been considered a relatively rare, densely clustered, posttranslational modification (PTM) occurring in mucins and mucin-like proteins. Such O-glycosylated regions are thought to attract water molecules, stabilize protein structure, extend the polypeptide backbone, and protect from proteolytic cleavage.7 Recent glycoproteomics studies in cell lines, however, indicate that, far from being specific to mucin-like regions, O-glycosylation is a ubiquitous PTM and >80% of proteins trafficking through the secretory pathway are estimated to be O-glycosylated.1,8 Furthermore, it is likely that O-glycans play a role in diverse physiologic systems because altered O-glycosylation is associated with IgA nephropathy,9 Tn syndrome,10 Crohn disease,11 tumorigenesis,12 impaired leukocyte recruitment,13 and high-density lipoprotein levels.14 Dissection of the molecular mechanisms by which O-glycans affect these systems is, however, currently impeded by a lack of knowledge of specific O-glycan sites in vivo.
Major O-glycan structures in human serum and endothelial cells. The majority of O-glycans in human serum4 and HUVECs5 are formed from sialylated Core 1 (Galβ1-3GalNAcαSer/Thr) and Core 2 (GlcNAcβ1-6[Galβ1-3] GalNAcαSer/Thr) structures. Mature glycans are capped with variable numbers of negatively charged sialic acid (Neu5Ac), which can be removed enzymatically by neuraminidases. O-glycans can also be found in fucosylated and sulfated forms. An example of a complex Core 2 O-glycan carrying a 6-O-sulfated sialyl-Lewis x terminal structure is shown at right. The lectin enrichment strategy employed in this study uses VVA and PNA to predominantly target the biosynthetic intermediate, Tn, and Core 1 O-glycans.
Major O-glycan structures in human serum and endothelial cells. The majority of O-glycans in human serum4 and HUVECs5 are formed from sialylated Core 1 (Galβ1-3GalNAcαSer/Thr) and Core 2 (GlcNAcβ1-6[Galβ1-3] GalNAcαSer/Thr) structures. Mature glycans are capped with variable numbers of negatively charged sialic acid (Neu5Ac), which can be removed enzymatically by neuraminidases. O-glycans can also be found in fucosylated and sulfated forms. An example of a complex Core 2 O-glycan carrying a 6-O-sulfated sialyl-Lewis x terminal structure is shown at right. The lectin enrichment strategy employed in this study uses VVA and PNA to predominantly target the biosynthetic intermediate, Tn, and Core 1 O-glycans.
The hemostatic system comprises platelet aggregation, coagulation, and fibrinolysis and is critical for the maintenance of vascular integrity. Within the hemostatic system, the presence of glycans on individual proteins has been demonstrated to alter expression, clearance, and catalytic activity.15 O-glycans on von Willebrand factor (VWF) have been associated with changes in VWF plasma concentration,16 platelet binding,17 and response to shear stress.18 O-glycosylation is also required for P-selectin–dependent leukocyte rolling,19,20 and O-glycosylation of platelet glycoprotein Ib α (GP1BA) is important for binding to VWF.21 Similarly, coagulation factors have been reported to be modified by O-glycosylation. Activation of coagulation factor X (FX) by Russell’s viper venom and Xase is altered in the presence of O-glycans,22 as is the sensitivity of coagulation factor XII (FXII) to contact activation, with loss of FXII O-glycans recently implicated in the pathogenesis of hereditary angioedema.23 Furthermore, murine studies have shown that O-glycans are important for the hemostatic process in vivo, because truncation of O-glycans results in severe bleeding deficits, aberrant angiogenesis, and platelet biogenesis, whereas loss of specific O-glycan sites leads to increased bleeding time and VWF insufficiency.24-26
These examples suggest important functions of O-glycosylation, yet, primarily because of technical limitations, we have little information on where the O-glycans are localized. Unlike N-linked glycosylation, no consensus sequence has been found for O-linked glycosylation, which further complicates analysis and prediction of O-glycan sites. Recent developments in mass spectrometry and glycan-enrichment methods, however, have enabled global studies of site-specific glycosylation using native tissues.8,27-30 Such glycoproteomic analyses have been applied to identify O-glycosites in human cerebrospinal fluid28 and urine.29 More recently, substantial progress has been made in the analysis of human plasma and serum,30,31 although the overall number of glycopeptides identified has been modest because of difficulties achieving high-sensitivity glycopeptide enrichment.
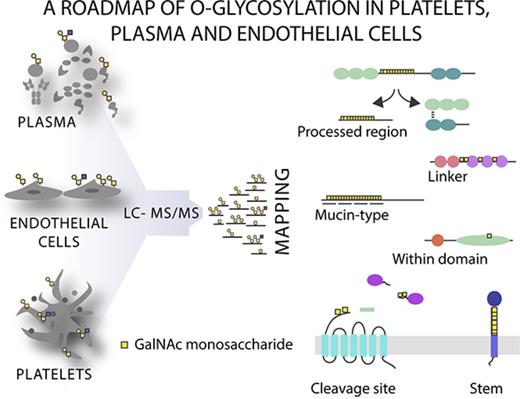
Here we extend this knowledge using a dual vicia villosa agglutinin (VVA) and peanut agglutinin (PNA) lectin weak affinity chromatography (LWAC) enrichment strategy1,8,32 coupled to higher-energy collisional dissociation (HCD) and electron transfer dissociation (ETD) liquid chromatography mass spectrometry (LC-MS) and present the first large-scale identification of O-glycosites in platelets, plasma, and endothelial cells.
Materials and methods
For a more detailed methods section see the supplemental Materials.
Glycoproteomic analysis
AB RhD–positive platelets from 4 random donors and plasma were obtained from the Blood Bank of the Capital Region and harvested according to standard protocols. Primary Human Umbilical Vein Endothelial Cells (HUVEC) were purchased from Life Technologies. 0.5 mL of packed cells or 2 mL plasma was prepared for O-glycoproteomic analysis as previously described.33,34 Briefly, samples were sonicated, de-sialylated with 0.1 U/mL Clostridium perfringens neuraminidase Type VI (Sigma) for 1 hour at 37°C, reduced (5 mM dithiothreitol, 60°C, 30 minutes), alkylated (10 mM iodoacetamide, room temperature, 30 minutes), and subjected to overnight digestion with MS-grade trypsin (Roche) or chymotrypsin (Roche) at 1:50 wt/wt. Samples were then purified on a C18 SepPak (Waters) and dried by SpeedVac. Glycopeptides were enriched by lectin chromatography using PNA- or VVA-agarose (VectorLabs) on an AKTA fast protein liquid chromatography system. Eluted glycopeptides were then de-salted using C18 Stage Tips before Orbitrap LC-MS/MS analysis. MS/MS spectra were interrogated against the nonredundant human proteome using the SEQUEST-HT search engine in Proteome Discoverer. These data can be fully interrogated online (http://glycodomain.glycomics.ku.dk/doi/10.1182/bloodadvances.2016002121/) using the Glycodomain Viewer,1 and mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE35 partner repository with the dataset identifier PXD004590.
Bioinformatics
All bioinformatics and statistical analyses were performed using R statistical software. Where required, data were compared with a background hemostatic proteome.36-38 Data sets used during the analysis are listed in supplemental Table 1. Proportional Venn Diagrams were generated using BioVenn.39 GO term analysis was performed using the DAVID bioinformatics resource (https://david.ncifcrf.gov/).40 Only GO terms demonstrating a twofold or greater change and represented by ≥10 proteins with a PBenjamini < .001 were considered.
Synthesis of glycopeptide substrates
Synthetic 20-mer peptides (NEO biolabs) were designed around known glycosylation sites and subjected to in vitro glycosylation using recombinant glycosyltransferases expressed as soluble secreted truncated proteins in insect cells, and were purified as described previously.41 10 µg of acceptor peptides and 2 mM uridine triphosphate–GalNAc as the sugar donor were prepared in 25 µL buffer containing 25 mM cacodylic acid sodium pH 7.4, 10 mM MnCl2, 0.25% Triton X-100. After 4- and 16-hour incubations at 37°C, reaction products were analyzed by MALDI-TOF-MS to determine glycosyltransferase specificity. For cleavage assays, 50 μg of peptide was glycosylated as described before, acidified, and purified by ultra-high-performance liquid chromatography on a C18 column. Control peptides were subjected to the same process in the absence of donor sugar.
In vitro cleavage assays
1.25 nmol (100 μM) glycosylated or control peptides were incubated for the indicated times with 50 nM plasmin (Sigma) or 50 nM human thrombin (Sigma) in 100 mM Tris-HCl, pH 7.4, 100 mM NaCl; 500 nM coagulation factor Xa (Haematologic Technologies) in 40 mM Tris-HCl, pH 8, 100 mM NaCl, 2 mM CaCl2; 50 nM neutrophil elastase (ENZO Life Science) in 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), pH 7.4, 150 mM NaCl, 0.05% Brij-35; 50 nM MMP7 or 100 nM MMP12 (R&D Systems) in 50 mM Tris-HCl, pH 7.4, 10 mM CaCl2, 150 mM NaCl, 0.05% Brij-35, 0.01 mM ZnCl2. Both MMP7 and MMP12 were activated with 1 mM AMPA for 30 minutes at 37°C before assay.
Results
Human platelet, plasma, and endothelial cell proteins are extensively O-glycosylated
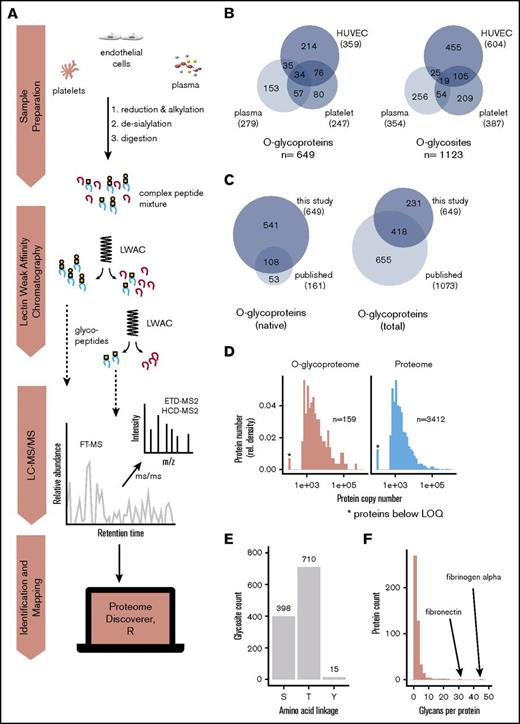
To identify which proteins are O-glycosylated and where the O-glycans are localized, we used our 2-step lectin enrichment strategy coupled to LC-MS/MS to analyze the global native O-glycoproteome. Enrichment of glycopeptides is essential for glycan detection in complex samples, and the strategy to identify O-glycosylation sites is shown in Figure 2A. We first performed lectin profiling of human platelets, plasma, and endothelial cells and demonstrated that all samples predominantly expressed sialylated Core 1 O-glycan structures (sialyl-T; supplemental Figure 1). A small amount of the nonsialylated Core 1 structure (T) was detected in nontreated endothelial cells, but not in platelet or plasma samples. Endothelial and, to a much lower degree, platelet samples also demonstrated weak expression of the nonsialylated biosynthetic intermediate, Tn, on a small subset of proteins. To identify specific sites of O-glycosylation, de-sialylated samples were enriched by LWAC using PNA and VVA lectins, which recognize T and Tn structures, respectively. Glycopeptides were subsequently identified using HCD and ETD LC-MS/MS (Figure 2A; supplemental Tables 2-5). Using this approach, a total of 649 unique O-glycoproteins were detected (Figure 2B). On these glycoproteins, 1123 O-glycosites could be unambiguously assigned. A further 547 ambiguous O-glycopeptides carrying 700 glycans were identified in which the glycan position could not be confidently assigned within the peptide because of insufficient MS2 fragmentation to report the corresponding diagnostic fragment ions or missing ETD spectra. In total, 1848 O-glycans were identified. These data represent a substantial increase in the existing knowledge (Figure 2C). Because mass spectrometric analysis of native samples such as platelet lysates is challenging due to the saturation of signals by highly abundant proteins, we compared the dynamic range of the platelet O-glycoproteome to the published quantitative proteome36 to better understand the depth of coverage achieved. Even in the absence of extensive precolumn separation, we found the range of glycoprotein detection to recapitulate that of total protein expression. With glycopeptide enrichment, low copy number (<500) proteins were detected, demonstrating that the method can be applied to complex native samples (Figure 2D).
Enrichment and identification of glycopeptides in the hemostatic system. (A) Depiction of the proteomics workflow. Human platelet, plasma, and endothelial samples were reduced, alkylated, de-sialylated using neuraminidase, and subsequently digested using either chymotrypsin or trypsin. Glycopeptides were then enriched from the resulting complex peptide mixtures by sequential LWAC using VVA and PNA lectins. VVA enrichment was not used for plasma samples because of the absence of Tn glycans; however, plasma samples were further separated using isoelectric focusing to reduce sample complexity. Fractions containing glycopeptides were separated by online reverse-phase liquid chromatography followed by identification using Orbitrap FTMS. (B) Glycoproteins and glycosites identified in each sample in this study. (C) Overlap of the O-glycoproteins identified in this study with previously published O-glycoproteins. Left, O-glycoproteins identified in native samples. Right, overlap with all reported O-glycoproteins. (D) Sensitivity of LWAC detection. The abundance of each platelet O-glycoprotein was determined based on the platelet protein copy numbers reported in Burkhart et al36 and plotted as a histogram (left) alongside the total platelet protein abundance obtained from the same study (right). Proteins with <500 copies are below the limit of quantification and are indicated by an asterisk. Comparison of the 2 distributions indicates that O-glycoproteins are detected over the full dynamic range of protein expression and include a similar proportion of proteins that are present below the limit of quantification. Note that membrane proteins are not quantified and therefore excluded from analysis. (E) Count of Ser-, Thr-, and Tyr-linked O-glycans. (F) Number of unambiguous glycosites per protein.
Enrichment and identification of glycopeptides in the hemostatic system. (A) Depiction of the proteomics workflow. Human platelet, plasma, and endothelial samples were reduced, alkylated, de-sialylated using neuraminidase, and subsequently digested using either chymotrypsin or trypsin. Glycopeptides were then enriched from the resulting complex peptide mixtures by sequential LWAC using VVA and PNA lectins. VVA enrichment was not used for plasma samples because of the absence of Tn glycans; however, plasma samples were further separated using isoelectric focusing to reduce sample complexity. Fractions containing glycopeptides were separated by online reverse-phase liquid chromatography followed by identification using Orbitrap FTMS. (B) Glycoproteins and glycosites identified in each sample in this study. (C) Overlap of the O-glycoproteins identified in this study with previously published O-glycoproteins. Left, O-glycoproteins identified in native samples. Right, overlap with all reported O-glycoproteins. (D) Sensitivity of LWAC detection. The abundance of each platelet O-glycoprotein was determined based on the platelet protein copy numbers reported in Burkhart et al36 and plotted as a histogram (left) alongside the total platelet protein abundance obtained from the same study (right). Proteins with <500 copies are below the limit of quantification and are indicated by an asterisk. Comparison of the 2 distributions indicates that O-glycoproteins are detected over the full dynamic range of protein expression and include a similar proportion of proteins that are present below the limit of quantification. Note that membrane proteins are not quantified and therefore excluded from analysis. (E) Count of Ser-, Thr-, and Tyr-linked O-glycans. (F) Number of unambiguous glycosites per protein.
We next sought to define the general properties of native O-glycosylation using a bioinformatic analysis of the location, distribution, and clustering of identified O-glycosites. As previously described in cell studies,1 we found Thr is preferentially glycosylated over Ser, whereas Tyr is only rarely glycosylated (Figure 2E). In contrast to the canonical view of O-glycosylation as a high-density mucin-type modification, but in line with previous findings in cell lines,1 76% of glycoproteins identified in the present study carried ≤3 sites of O-glycosylation (Figure 2F). Only a small proportion (28 proteins, 4.3%) were found to be highly glycosylated (>10 sites), with fibrinogen α and fibronectin the most highly glycosylated proteins identified with >30 sites each. Glycosylation was also found to be heterogeneous because analysis of overlapping, multisite peptides indicated that O-glycosylation often occurred at substoichiometric levels (supplemental Figure 2A). Similarly, although the majority of glycans identified were T rather than Tn structures (supplemental Figure 2B), there was considerable structural heterogeneity at individual sites (supplemental Figure 2C). Interestingly, when analyzing the distribution of identified glycan structures, proteins carrying solely Tn-glycans were highly enriched for the annotation endoplasmatic reticulum, potentially indicative of a discrete functional role for these glycoproteins (supplemental Figure 2B, inset).
Previously it has been reported that GP1BA,42 P-selectin glycoprotein ligand 1 (PSGL1)43,44 and VWF,45 among others, carry Core 2 structures and that these structures make up ∼2% of the plasma O-glycoproteome.5 Enrichment and MS analysis of these structures is problematic, and only a handful of sites carrying complex O-glycans have been identified using glycoproteomics.29,46,47 We reasoned, however, that a subset of these sites may be detectable because of co-elution with T/Tn peptides in our study. We therefore developed a script to search for spectra containing [HexNAc2]+ (m/z 407) and [HexHex-NAc2]+ (m/z 569) diagnostic peaks to determine whether there was evidence of branched O-glycans in our data set. In addition, we applied the algorithm described by Halim et al to predict the presence of GlcNAc monosaccharides.48 Combining these data, we found spectra representing branched, possible core 2, structures on 64 (9.9%) proteins. Glycosites predicted to carry these structures are highlighted in supplemental Tables 2-5 and supplemental Figure 3.
The majority of O-glycosites discovered to date have been identified in immortalized cell lines that have been glycoengineered to homogeneously express the truncated Tn antigen.1 Because the effects of O-glycan truncation on O-glycan location are unknown, it is not clear whether previously identified sites are representative of native glycosylation. We therefore compared sites identified here with previously published sites to determine whether general properties of O-glycosylation were conserved between glycoengineered cells and native samples. No substantial differences were noted. Glycosites were found to reside in disordered regions within the extracellular or lumenal segments of proteins (supplemental Figure 4A). The majority of glycans occurred as individual sites as previously described49 ; however, where multiple sites were identified, they were found to weakly cluster (supplemental Figure 4B). No clear consensus sequence could be defined for O-glycans, although the amino acids Pro, Ser, and Thr were over-represented, and negatively charged amino acids were under-represented around glycosylation sites relative to randomly sampled Ser/Thr (supplemental Figure 4C). Glycosites were found along the whole length of proteins, with a small but significant enrichment in the N-terminal (at 40-70 amino acids; supplemental Figure 5). Taken together, these data demonstrate that O-glycosylation is a ubiquitous modification of disordered regions in the extracellular space. Furthermore, the site distribution and localization properties between hemostatic native tissues and immortalized cells are conserved.
Site-specific O-glycosylation of key hemostatic proteins
One of the central functions of proteins from platelets, plasma, and endothelial cells is the maintenance of vascular integrity accomplished through regulation of platelet activation, blood clotting, fibrinolysis, and vascular repair. Together, these processes are referred to as hemostasis. Key hemostatic proteins are major therapeutic targets and, as such, site-specific O-glycosylation has been thoroughly investigated on many of these proteins. We therefore compared O-glycosites identified on key hemostatic proteins in this study with previously reported sites to (1) highlight novel findings, (2) provide an updated summary of known O-glycosites for the field, and (3) illustrate overall patterns of O-glycosylation within protein families and across multiple protein classes (Figures 3 and 4; supplemental Figure 6).50,51 For many sites, this is the first evidence of native (in vivo) O-glycosylation of these proteins (supplemental Figure 6). Examples of this can be seen on Protein S, prothrombin, urokinase, carboxypeptidase B2, and plasminogen activator inhibitor-1, among others (Figure 3; supplemental Figure 6). Similarly, although the platelet receptor glycoprotein (GP)Ib-IX-V complex is known to be O-glycosylated, only a single probable O-glycosite (Thr308 on GP1BA) has been described.51 Here we were able to identify O-glycosites on all members of the complex (Figure 4). Novel O-glycosites were also discovered on integrin receptors, with the occurrence of O-glycosites within the VWFA domain of β-integrins being of particular note. The most conspicuous finding, however, was the extensive glycosylation of coagulation factor V (FV), fibrinogen α, and fibronectin, which far exceeded prior expectations.
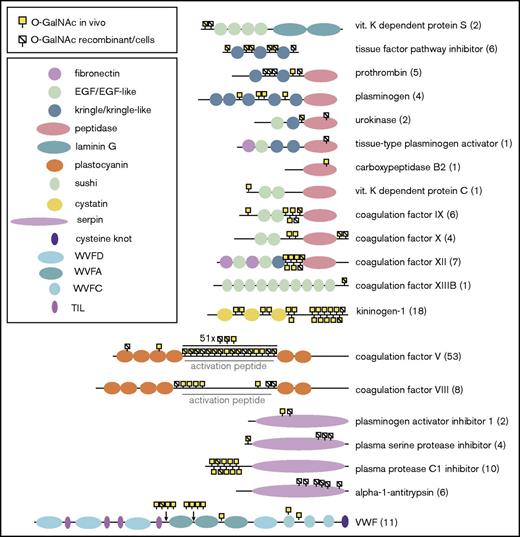
Summary of total O-glycosylation of coagulation factors and inhibitors. Protein schematics illustrating the location of O-glycosites relative to Uniprot domains on coagulation factors and inhibitors. The total number of unambiguous sites is indicated in brackets on the right. Sites identified in native (“in vivo”) samples in either this study or prior publications are indicated by a yellow square. Sites identified on recombinant proteins or in immortalized cell lines are indicated by a stroke. Many sites are located at the N-termini of the protein, or within processed regions (either propeptides, regions cleaved for activation, or on activation peptides). This is exemplified by the extensive glycosylation of the activation peptide of FV, but is also evident on the majority of proteins shown above. EGF, epidermal growth factor; TIL, trypsin inhibitor-like.
Summary of total O-glycosylation of coagulation factors and inhibitors. Protein schematics illustrating the location of O-glycosites relative to Uniprot domains on coagulation factors and inhibitors. The total number of unambiguous sites is indicated in brackets on the right. Sites identified in native (“in vivo”) samples in either this study or prior publications are indicated by a yellow square. Sites identified on recombinant proteins or in immortalized cell lines are indicated by a stroke. Many sites are located at the N-termini of the protein, or within processed regions (either propeptides, regions cleaved for activation, or on activation peptides). This is exemplified by the extensive glycosylation of the activation peptide of FV, but is also evident on the majority of proteins shown above. EGF, epidermal growth factor; TIL, trypsin inhibitor-like.
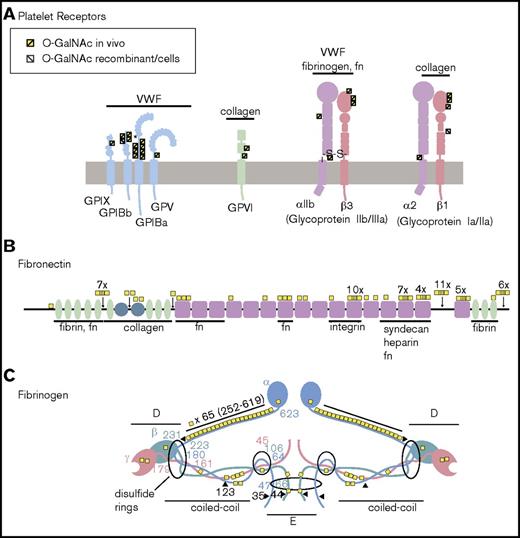
O-glycosylation of platelet receptors, fibronectin, and fibrinogen. (A) Platelet receptors. Platelet receptors are critical mediators of platelet activation and adherence, and include the well-described GPIB-V-IX complex, the collagen binding GPVI, and multiple integrin receptors.50 GPIB α is part of the GPIB-V-IX (VWF receptor) complex and contains a mucin-like macroglycopeptide stem region, which is known to be O-glycosylated; however, the specific sites are poorly described, with only a single probable site at Thr308 (indicated by and asterisk).51 In this study, 7 glycosites were identified in this region. Novel O-glycosylation was identified on all other members of the complex in regions flanking the leucine-rich repeats. An additional novel O-glycosite was also detected on the collagen receptor GPVI. Integrin receptors (glycoproteins IIb/IIIa and Ia/IIa) were also found to be O-glycosylated at multiple sites. In particular, novel glycosylation was identified in the VWFA domains of β integrins (β1 and β3), and a novel glycosite was identified juxtaposed to the transmembrane region of integrin α2. Major ligands for each of the receptors are indicated above the receptors. (B) Fibronectin. Four sites of O-glycosylation have been described for fibronectin, 3 sites located in the variable region (T2024, T2064, T2065) and a fourth site in an N-terminal linker region (T279). All sites except T2024 were identified in this study. Moreover, in total, hemostatic fibronectin was found to carry 31 unambiguous glycosites and 14 additional ambiguous sites of glycosylation. In total, 71 unambiguous sites have been identified on fibronectin from all sources. (C) Fibrinogen. Fibrinogen α was found to be extensively glycosylated, with 45 glycosites identified in hemostatic samples and 68 sites identified across all sources. The glycosites were distributed across the protein, with the majority located within the coiled-coiled domain. Fibrinogen β and γ were much less glycosylated and were found to carry just 2 and 1 sites in this study, respectively. In total, across all studies, 4 sites have been detected on fibrinogen β and 7 on fibrinogen γ. Because of the large number of identified sites, no distinction is made between native (in vivo) and in vitro glycosylation on fibronectin and fibrinogen. Fibronectin (fn).
O-glycosylation of platelet receptors, fibronectin, and fibrinogen. (A) Platelet receptors. Platelet receptors are critical mediators of platelet activation and adherence, and include the well-described GPIB-V-IX complex, the collagen binding GPVI, and multiple integrin receptors.50 GPIB α is part of the GPIB-V-IX (VWF receptor) complex and contains a mucin-like macroglycopeptide stem region, which is known to be O-glycosylated; however, the specific sites are poorly described, with only a single probable site at Thr308 (indicated by and asterisk).51 In this study, 7 glycosites were identified in this region. Novel O-glycosylation was identified on all other members of the complex in regions flanking the leucine-rich repeats. An additional novel O-glycosite was also detected on the collagen receptor GPVI. Integrin receptors (glycoproteins IIb/IIIa and Ia/IIa) were also found to be O-glycosylated at multiple sites. In particular, novel glycosylation was identified in the VWFA domains of β integrins (β1 and β3), and a novel glycosite was identified juxtaposed to the transmembrane region of integrin α2. Major ligands for each of the receptors are indicated above the receptors. (B) Fibronectin. Four sites of O-glycosylation have been described for fibronectin, 3 sites located in the variable region (T2024, T2064, T2065) and a fourth site in an N-terminal linker region (T279). All sites except T2024 were identified in this study. Moreover, in total, hemostatic fibronectin was found to carry 31 unambiguous glycosites and 14 additional ambiguous sites of glycosylation. In total, 71 unambiguous sites have been identified on fibronectin from all sources. (C) Fibrinogen. Fibrinogen α was found to be extensively glycosylated, with 45 glycosites identified in hemostatic samples and 68 sites identified across all sources. The glycosites were distributed across the protein, with the majority located within the coiled-coiled domain. Fibrinogen β and γ were much less glycosylated and were found to carry just 2 and 1 sites in this study, respectively. In total, across all studies, 4 sites have been detected on fibrinogen β and 7 on fibrinogen γ. Because of the large number of identified sites, no distinction is made between native (in vivo) and in vitro glycosylation on fibronectin and fibrinogen. Fibronectin (fn).
O-glycosylation occurs in a cell-specific manner in the hemostatic system
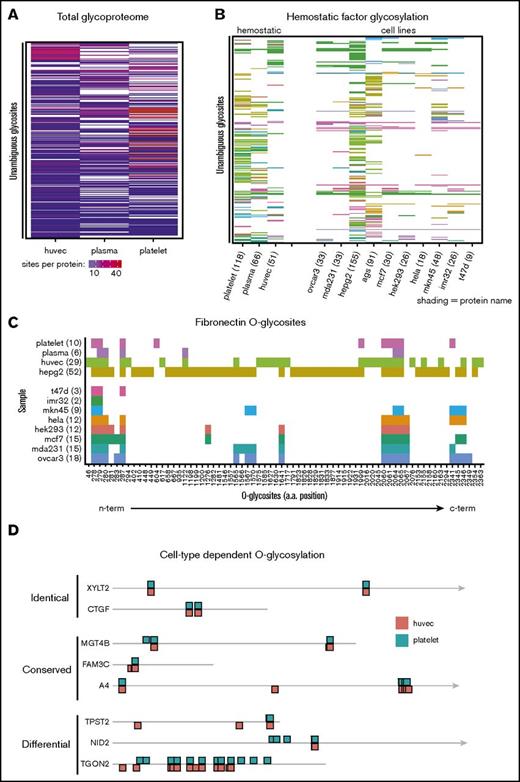
We next asked if proteins from different cellular sources were differentially O-glycosylated. Cells and tissues express distinct repertoires of GalNAc transferases (supplemental Figure 7) that can potentially fine-tune protein function in a cell-specific manner.52,53 Comparing the total O-glycoproteomes of each cellular source revealed that platelets, endothelial cells, and plasma proteins each express a unique O-glycoproteome (Figure 5A). This cell-specific behavior was found to be more pronounced when compared across a broader selection of cell types (Figure 5B). Such differences could, however, result from differential protein expression or GalNAc-T activity. Therefore, we analyzed the cell-specific glycosylation of fibronectin, which has been identified in multiple cell types (Figure 5C). We found 2 regions of glycosylation to be conserved (amino acid positions 278-287 and 2060-2067), but also observed multiple regions of cell-specific glycosylation. To ensure similar coverage between samples, we next analyzed the cell-specific glycosylation of proteins with ≥5 spectra in each sample and with congruent spectral counts. Again we found evidence for sample-specific glycosylation in parallel with conserved glycosylation patterns (Figure 5D). These findings open the possibility of cell-specific regulation of hemostatic proteins by cell-specific differential O-glycosylation.
Cells express unique O-glycoproteomes. To visualize the O-glycoproteome of each cell, the presence or absence of individual glycosites was illustrated using unclustered heat maps. Glycosites were ordered alphabetically by Uniprot accession and subsequently by position in the protein. (A) Total O-glycosylation illustrating the overlap of unique, unambiguous glycosites identified in each sample. (B) Comparison of O-glycosite identification on key hemostatic factors between different sample types. (C) A detailed view of differential O-glycosylation of fibronectin across samples. Individual O-glycosites identified in fibronectin are plotted by sample type in order of position on the protein, with N-terminal sites plotted closest to the origin. Numbers indicate total glycosite count per protein. (D) Differential O-glycosylation of proteins in the hemostatic system. The hemostatic O-glycoproteome was filtered to identify proteins represented by >5 spectra in each sample and with similar spectral counts between samples (total protein spectral counts differing by <10) to ensure comparable O-glycosite coverage was achieved between samples. These proteins demonstrated both overlapping and also cell-specific glycosylation as exemplified in (D). Drawing is to scale; the arrowhead indicates the protein extends beyond illustration. Shading indicates number of sites per protein (A), protein identity (B), or sample type (C). a.a., amino acid.
Cells express unique O-glycoproteomes. To visualize the O-glycoproteome of each cell, the presence or absence of individual glycosites was illustrated using unclustered heat maps. Glycosites were ordered alphabetically by Uniprot accession and subsequently by position in the protein. (A) Total O-glycosylation illustrating the overlap of unique, unambiguous glycosites identified in each sample. (B) Comparison of O-glycosite identification on key hemostatic factors between different sample types. (C) A detailed view of differential O-glycosylation of fibronectin across samples. Individual O-glycosites identified in fibronectin are plotted by sample type in order of position on the protein, with N-terminal sites plotted closest to the origin. Numbers indicate total glycosite count per protein. (D) Differential O-glycosylation of proteins in the hemostatic system. The hemostatic O-glycoproteome was filtered to identify proteins represented by >5 spectra in each sample and with similar spectral counts between samples (total protein spectral counts differing by <10) to ensure comparable O-glycosite coverage was achieved between samples. These proteins demonstrated both overlapping and also cell-specific glycosylation as exemplified in (D). Drawing is to scale; the arrowhead indicates the protein extends beyond illustration. Shading indicates number of sites per protein (A), protein identity (B), or sample type (C). a.a., amino acid.
An unbiased screen to identify protein features colocalizing with O-glycosylation
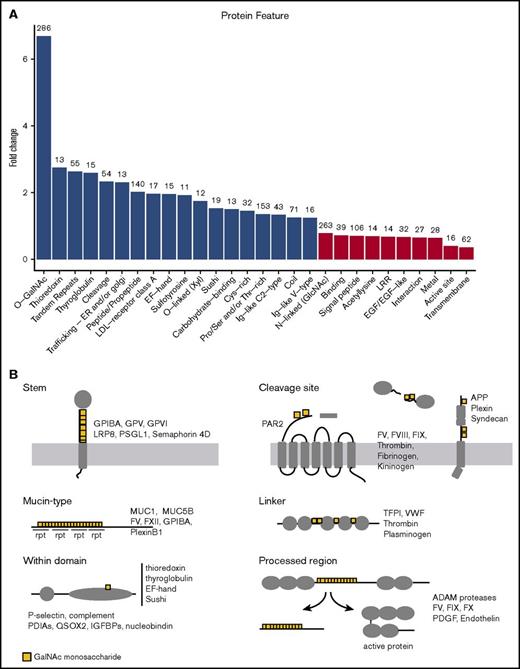
The relative location of a PTM can be highly informative of its potential function. For example, acetylation at a protein N-terminal will likely alter the protein’s stability, whereas the same modification on an active-site lysine may modify catalytic activity. We noted during our manual analysis that glycosites were repeatedly found near certain protein annotations such as phosphorylation sites, proteolytic cleavage sites, serpin reactive center loops, and in helix/coiled regions, VWF domains, and protein stems/linkers. These subpatterns could suggest discrete roles for O-glycans worthy of further investigation; however, such manual analysis is biased by the types of proteins studied, prior knowledge, and potential effects of nonrandom Ser/Thr distribution. Therefore, we performed a systematic and unbiased association study using the full complement of protein annotations available from UniprotKB54 and all O-glycosites described to date1,8,28-30,32,47,55-57 (supplemental Table 1). This analysis identifies whether O-glycosites are over- or underenriched around (± 15 amino acids) each protein annotation relative to the expected frequency based on the background distribution of Ser/Thr residues, and is somewhat analogous to a gene ontology study, in which gene ontologies are replaced with protein annotations (Figure 6A). Among the most significant findings, individual O-glycosites were associated with cleavage sites in accordance with previous data on proprotein convertase and metalloprotease cleavage.58,59 We also observed association with propeptides and bioactive peptides, and further inspection indicated that glycosites were localized within the processed region, rather than around the periphery. Although rare, O-glycosites were associated with a small group of protein domains including the low-density lipoprotein receptor class A, thioredoxin, EF-hand, Ig-like, and sushi domains. Signal and transmembrane domains were under-represented as expected. Interestingly, N-glycosites, nucleic acid binding regions, and binding regions in general were less frequently associated with an O-glycosite than expected. Although the data were supported by few sites, it was notable that glycosites were also found in proximity to regions involved in endoplasmic reticulum/Golgi trafficking and sulfotyrosine annotations. Identified subtypes of O-glycosylation including selected examples are shown in Figure 6B.
Enrichment of protein annotations around O-glycosites. (A) Protein annotations from the UniprotKB database were used to determine whether specific protein features were enriched around (± 15 amino acids) O-glycosites relative to the background Ser/Thr distribution. Annotated features with a >1.2 fold-change and P < .01 (Fisher’s exact test, Bonferroni correction) are illustrated on the graph above. Note that O-glycosites often occurred in the vicinity of Ser/Thr phospho-sites and disulfide-bonded Cys, but at the expected frequency. Numbers above the bars indicate the count of O-glycosites found in the vicinity of the annotation. Fold-change indicates the under- or overenrichment of O-glycosites relative to the background frequency of Ser/Thr residues. Only annotations represented by >5 proteins and 10 glycosites were included in the analysis. (B) Graphic depiction showing examples of the different types of O-glycosylation identified in this study. rpt, repeat.
Enrichment of protein annotations around O-glycosites. (A) Protein annotations from the UniprotKB database were used to determine whether specific protein features were enriched around (± 15 amino acids) O-glycosites relative to the background Ser/Thr distribution. Annotated features with a >1.2 fold-change and P < .01 (Fisher’s exact test, Bonferroni correction) are illustrated on the graph above. Note that O-glycosites often occurred in the vicinity of Ser/Thr phospho-sites and disulfide-bonded Cys, but at the expected frequency. Numbers above the bars indicate the count of O-glycosites found in the vicinity of the annotation. Fold-change indicates the under- or overenrichment of O-glycosites relative to the background frequency of Ser/Thr residues. Only annotations represented by >5 proteins and 10 glycosites were included in the analysis. (B) Graphic depiction showing examples of the different types of O-glycosylation identified in this study. rpt, repeat.
O-glycosylation regulates cleavage of hemostatic factors
From our analysis, it was apparent that a number of O-glycosites were closely juxtaposed with cleavage sites critical to the regulation of hemostasis. Therefore, we investigated whether O-glycans could regulate hemostatic factor processing in vitro using synthetic 20-mer peptides. We first tested whether identified glycosites could be glycosylated in vitro using the 3 major glycosyltransferases: GalNAc-T1, T2, and T3. We found that 31 of the 33 peptides tested could be glycosylated by ≥1 enzyme in overnight reactions, with a substantial overlap in enzyme specificity (supplemental Figure 8). We then selected 8 of the glycosylated peptides, which also contained a reported cleavage site, and investigated whether the presence of O-glycans altered the cleavage response in an in vitro time-course assay (Figure 7). We found that O-glycosylation partially inhibited the cleavage response to neutrophil elastase, thrombin, and MMP12 in a glycosite- and glycoform-specific manner, with a marked inhibition in the cleavage of protease-activated receptor 2 (PAR2) and tissue factor pathway inhibitor 1 (TFPI1) by neutrophil elastase and the cleavage of protein S by thrombin. These findings indicate that O-glycosylation is able to alter the sensitivity toward proteolytic cleavage across multiple protein- and protease-families in the hemostatic system.
Glycosylation inhibits proteolytic processing. In vitro cleavage analysis of glycopeptides and their nonglycosylated counterparts. Synthetic peptides were in vitro–glycosylated and then subjected to a protease digestion time course with the indicated enzymes. (A) Summary of results. Identified cleavage sites are indicated by an arrow. Proteases affected by glycosylation are colored blue (partial inhibition) or red (complete inhibition). Glycosites identified by prior glycoproteomic analysis are shown in red; glycosites identified by LC-MS/MS on in vitro glycosylated peptides are indicated by a yellow box. Glycosylation of the TFPI1_2 peptide both delayed and repositioned NE cleavage. (B) Example matrix-assisted laser desorption/ionization–time-of-flight spectra showing glycoform-dependent inhibition of thrombin serine protease activity. The monoisotopic mass of the cleavage product is indicated in red. NE, neutrophil elastase; THRB, thrombin; PLMN, Plasmin. n = 2.
Glycosylation inhibits proteolytic processing. In vitro cleavage analysis of glycopeptides and their nonglycosylated counterparts. Synthetic peptides were in vitro–glycosylated and then subjected to a protease digestion time course with the indicated enzymes. (A) Summary of results. Identified cleavage sites are indicated by an arrow. Proteases affected by glycosylation are colored blue (partial inhibition) or red (complete inhibition). Glycosites identified by prior glycoproteomic analysis are shown in red; glycosites identified by LC-MS/MS on in vitro glycosylated peptides are indicated by a yellow box. Glycosylation of the TFPI1_2 peptide both delayed and repositioned NE cleavage. (B) Example matrix-assisted laser desorption/ionization–time-of-flight spectra showing glycoform-dependent inhibition of thrombin serine protease activity. The monoisotopic mass of the cleavage product is indicated in red. NE, neutrophil elastase; THRB, thrombin; PLMN, Plasmin. n = 2.
Discussion
Here we took advantage of recent progress in lectin enrichment and high-resolution MS to selectively identify site-specific O-GalNAc type glycosylation in platelets, plasma, and endothelial cells. With this approach, we were able to substantially increase our knowledge of the native human O-glycoproteome by unambiguously identifying 1123 sites on 649 proteins, including identification of 541 sites and 231 O-glycoproteins that had not previously been found in native samples.30,57,60 It should be noted, however, that these data likely underestimate the total number of O-glycans present because of the inherent limitations of shotgun proteomics.27 This is of particular concern in regards to mucin-like repeat regions and highly homologous protein families that are refractory to mapping by mass spectrometry. In such cases, the NetOGlyc 4.0 O-glycosite predictor can be used to complement experimental approaches, because probable mucin-like regions can be identified by regions of predicted high-density glycosylation, as exemplified by GP1BA42,61 and PSGL1.43,62
The identification of a large number of O-glycosites enabled investigation of the potential function of glycosylation by determining which annotated protein features colocalize with O-glycosites. Here, we assessed whether glycosites were over- or underenriched around protein features compared with the background distribution of Ser/Thr residues. As expected, we found glycosites to be enriched around tandem repeat or Pro/Ser/Thr rich regions, presumably indicative of canonical mucin-type glycosylation. Within the hemostatic system, the coagulation factors V and XII, GP1BA, PSGL1, and Plexin B1 were identified with glycosites in such regions. This form of dense mucin-type glycosylation is thought to confer lubricating properties,63 extend the polypeptide backbone, reduce flexibility, and provide resistance to proteases.7,64 For example, O-glycans are thought to extend GP1BA from the platelet surface, allowing optimal interaction with VWF under shear stress.21 The presence of a mucin-type domain in the soluble coagulation factors could conceivably protect the factors from degradation and premature activation in circulation, but such a role has not yet been described. We also found a strong association between O-glycosylation and proteolytic cleavage sites. Accumulating data suggest that this colocalization is not simply the result of a propensity for such PTMs to occur in similar regions, because loss of O-glycosites has been directly shown to inhibit or activate protein processing by proteases.58,59,65 In the context of the hemostatic system, sialylation and O-glycosylation of VWF alter sensitivity to cleavage in a protease-specific manner.66 Similarly, O-glycosylation regulates the rate of cleavage of coagulation factor X22 and LRP867 ; and loss of extended O-glycans results in degradation of platelet GP1BA in Cosmc null mice.25 In the present study, glycosites were found juxtaposed with critical processing sites in a large number of key hemostatic proteins (Figure 5B). Furthermore, these data were supported by in vitro assays demonstrating that cleavage of Protein S by thrombin and cleavage of PAR2 and TFPI1 by neutrophil elastase are inhibited by the presence of O-glycosylation. Such results suggest that O-glycosylation might be a major regulator of protein stability and processing in hemostasis.
In addition to mucin-type regions and proteolysis, we found O-glycosites within propeptides and bioactive-peptides, which are proteolytically removed from the mature protein during hemostasis. Glycosylation within these regions was common on hemostatic factors, particularly coagulation factors and collagens. The functional consequences of this type of O-glycosylation have not been elucidated yet; however, because the majority of O-glycans carry at least one negatively charged sialic acid, the release of these regions facilitates a rapid change in the charge density of the protein and thereby may have substantial effect on protein confirmation and binding. Additional associations with tyrosine sulfation and endoplasmic reticulum/Golgi trafficking were noted. Tyrosine sulfation contributes to protein-protein binding68 and is of interest because coordinated binding of a Core 2 O-glycan and sulfated tyrosines on the N-terminal of PSGL1 is critical to its recognition by P-selectin.19,20 Glycosites were found in proximity to sulfotyrosines in coagulation factors VIII, IX, and XII, and also vitronectin, GP1BA, and nidogen 1. Similarly, proximity to endoplasmic reticulum/Golgi trafficking signals is notable because O-glycosylation has been demonstrated to regulate secretion, suggesting a function in vesicular transport.69-71 Given that vesicular trafficking signals remain poorly annotated, it will be of interest to further investigate the potential role of O-glycosylation in protein trafficking experimentally.
The function of O-glycans is not solely dependent on the position of the glycan, but also, to a large extent, elongation and the addition of terminal structures. Such structures act as ligands for carbohydrate-binding proteins and are often altered in pathogenic states (eg, in response to inflammatory signaling72 and during metastasis73 ). It is not currently possible to simultaneously identify both glycan structure and glycosite for complex O-glycans. A number of glycopeptides identified in this study, however, were predicted to carry GlcNAc residues suggestive of elongated/branched structures. We were able to predict the presence of such branched structures on GP1BA at 490-494 in keeping with previous reports indicating that the majority of GP1BA O-glycans were of Core 2 structure.74,75 In addition to GP1BA, a number of key hemostatic factors, including GP1BB, GPIX, GPVI, fibronectin, coagulation factors V and XII, and P-selectin, carried branched structures suggesting that complex O-glycans (including blood group ligands) may be present on a substantial number of extracellular proteins. Notably, most sites identified carrying a branched structure were also identified with a simple Core 1 structure, indicating that there is variability in both site occupancy and glycan structure at individual sites.
In summary, O-glycosylation is a widespread modification of platelet, plasma, and endothelial cell proteins, occurring in disordered regions of extracellular proteins and presenting with variable occupancy and composition at individual sites. Although the precise role of O-glycans in hemostasis remains enigmatic, the distinct patterns of O-glycosylation identified here suggest the presence of multiple autonomous subgroups and provide a roadmap for future structure-function studies. Such information will be critical in the production of biotherapeutics and development of glycan-based biomarker studies.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Claus Ekstrom for statistical advice provided during the writing of this manuscript, Annika Lindkvist for assistance in the culture of HUVECs, and Simon Kuijpers for assistance with illustration.
This work was supported by The Danish Research Councils (Sapere Aude Research Leader grant [H.W.] and Talent Grant [K.T.S.]), The Mizutani Foundation, Kirsten og Freddy Johansen Fonden, A.P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, The Novo Nordisk Foundation, the Danish Strategic Research Council, the Lundbeck foundation, the program of excellence from the University of Copenhagen (CDO2016), and The Danish National Research Foundation (DNRF107).
Authorship
Contribution: S.L.K., H.J.J., K.T.S., S.Y.V., M.H.D., A.W., and H.H.W. designed experiments and wrote the paper; S.L.K., K.T.S., and T.D.M. performed experiments; and S.L.K., K.T.S., H.J.J., A.H., and S.Y.V. analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans H. Wandall, Department of Cellular and Molecular Medicine, Centre for Glycomics, University of Copenhagen, DK-2200 Copenhagen N, Denmark; e-mail: hhw@sund.ku.dk.

![Figure 1. Major O-glycan structures in human serum and endothelial cells. The majority of O-glycans in human serum4 and HUVECs5 are formed from sialylated Core 1 (Galβ1-3GalNAcαSer/Thr) and Core 2 (GlcNAcβ1-6[Galβ1-3] GalNAcαSer/Thr) structures. Mature glycans are capped with variable numbers of negatively charged sialic acid (Neu5Ac), which can be removed enzymatically by neuraminidases. O-glycans can also be found in fucosylated and sulfated forms. An example of a complex Core 2 O-glycan carrying a 6-O-sulfated sialyl-Lewis x terminal structure is shown at right. The lectin enrichment strategy employed in this study uses VVA and PNA to predominantly target the biosynthetic intermediate, Tn, and Core 1 O-glycans.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/7/10.1182_bloodadvances.2016002121/3/m_advances002121f1.jpeg?Expires=1767718319&Signature=UEKG5fohaYS6n7qiBPKU4~oiRnctOSY-n6AExwVELq9pYFHaKZdQ01pHTxQo7OUGiIlgH8cpp6Rdb7ago82qS1u6mZgXrGQvIr1gaQzrsM-4DFuOvmd3y~TCpPw~bmc7Q~on1oaBrWaQjPikurB9edV7ELsfGqsgN2MoCtrvPAOK5ckovHKDuBFwzxDqA8UvIcTPI2tpQfBlASZGgLNs-Uz9MMTNY4n9f6NW8qHzyWN-bhzl~aHnVkv-GdxMz6qaMvM7JK5eg7Dhywt1-Ook8b5tMR8KKHbdctPRN1ZbvuXiZ~VAQ3ZRdHB7Nm~XfdNCK~ZfTrcd1z5ZrM0ZS8qSKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)