Key Points
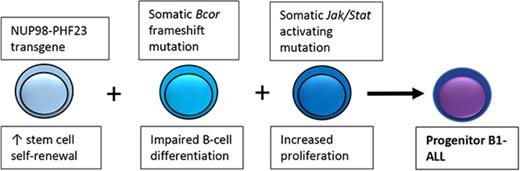
An NUP98-PHF23 fusion collaborates with acquired Bcor and Jak/Stat mutations to produce a pro–B-1 ALL.
Gene expression profile of murine pro–B-1 ALL resembles that of a subset of human ALL, suggesting some human ALLs arise from pro–B-1 B cells.
Abstract
B-1 and B-2 lymphocytes are derived from distinct developmental pathways and represent layered arms of the innate and adaptive immune systems, respectively. In contrast to a majority of murine B-cell malignancies, which stain positive with the B220 antibody, we discovered a novel form of B-cell leukemia in NUP98-PHF23 (NP23) transgenic mice. The immunophenotype (Lin− B220− CD19+ AA4.1+) was identical to that of progenitor (pro) B-1 cells, and VH gene usage was skewed toward 3′ V regions, similar to murine fetal liver B cells. Moreover, the gene expression profile of these leukemias was most similar to that of fetal liver pro-B fraction BC, a known source of B-1 B cells, further supporting a pro–B-1 origin of these leukemias. The NP23 pro–B-1 acute lymphoblastic leukemias (ALLs) acquired spontaneous mutations in both Bcor and Janus kinase (Jak) pathway (Jak1/2/3 and Stat5a) genes, supporting a hypothesis that mutations in 3 critical pathways (stem-cell self-renewal, B-cell differentiation, and cytokine signaling) collaborate to induce B-cell precursor (BCP) ALL. Finally, the thymic stromal lymphopoietin (Tslp) cytokine is required for murine B-1 development, and chromosomal rearrangements resulting in overexpression of the TSLP receptor (CRLF2) are present in some patients with high-risk BCP-ALL (referred to as CRLF2r ALL). Gene expression profiles of NP23 pro–B-1 ALL were more similar to that of CRLF2r ALL than non-CRLF2r ALL, and analysis of VH gene usage from patients with CRLF2r ALL demonstrated preferential usage of VH regions used by human B-1 B cells, leading to the suggestion that this subset of patients with BCP-ALL has a malignancy of B-1, rather than B-2, B-cell origin.
Introduction
A wide spectrum of B-cell malignancies has been recognized in humans. These malignancies are typically classified based on the presumed cell of origin and span the breadth of B-cell development, from immature B cells, such as progenitor (pro)–B cells, precursor (pre)–B cells, and pre-pro–B cells, to the more mature B cells, such as plasma cells. A variety of diagnostic tools, including clinical presentation, histology, immunophenotype, cytogenetics, and molecular genetics, have been used to characterize and classify these B-cell malignancies. However, even with these modern tools, a proportion of leukemias and lymphomas remain difficult to classify and are termed leukemias of ambiguous lineage or B-cell lymphoma, unclassifiable.1
Normal B-cell differentiation is thought to follow 1 of 2 developmental B-cell pathways, designated B-1 and B-2. B-2 B cells constitute the predominant class of B cells found in the spleen, lymph nodes, and peripheral blood and function in adaptive immunity (reviewed by Montecino-Rodriguez and Dorshkind2 ). B-1 B cells are rarely identified in the spleen or lymph nodes but instead predominate in the pleural and peritoneal cavities and are thought to represent an arm of the innate immune system. As such, they produce natural antibodies, which are not induced by exposure to foreign antigens, and typically recognize self-glycosylated and heavily glycosylated antigens. B-1 B cells have been more clearly defined and characterized in mice than in humans and are more prominent during fetal hematopoiesis than during adult hematopoiesis.2 A unique differentiation pathway for murine B-1 B cells has been characterized, with pro–B-1 cells in the murine fetal liver or bone marrow displaying a lineage negative (Lin−) B220 (CD45R)lo/− CD19+ AA4.1+ immunophenotype.3,4 Defining B-1 B cells in humans has been challenging, but human B-1 B cells have been described as CD3− CD19+ CD20+ CD27+ CD43+ CD69− CD70−, distinguishing them from naïve and memory B cells (CD43−), plasmablasts/plasma cells (CD19loCD20lo/−CD138±), CD43+-activated B cells (CD69++CD70++), and T cells (CD3+CD19−CD20−).5 In addition to their distinctive immunophenotype, umbilical cord B-1 B cells showed a skewed usage of VH chains, with preferential usage of VH3-30.5
Some mature B-cell malignancies, including cases of chronic lymphocytic leukemia in humans6-8 and CH lymphomas in mice,9 are suspected to arise from B-1 B cells, and expanded B-1 cell populations have been described in patients with systemic lupus erythematosus.10 Although B-1 lymphocytes can be transformed by transduction of a BCR-ABL retrovirus,11 leukemias of pro–B-1 B cells arising in genetically engineered mice have not been described. Herein we report that B-cell leukemias that arise in NUP98-PHF23 (NP23) transgenic mice12 originate from pro–B-1 B cells. Interestingly, these pro–B-1 B-cell leukemias (pro–B-1 ALLs) also acquire somatic mutations in Bcor and the Janus kinase (Jak) pathway; mutations in these genes have previously been implicated in acute myeloid leukemia (AML),13 myelodysplastic syndrome,14 and BCP-ALL.15,16
Materials and methods
Additional information on methods can be found in the supplemental Methods.
Mice
NP23 transgenic mice were generated as previously described.12 E2a:PBX1 and Eμ-RET samples were obtained from spleens of C57BL/6 or BALB/C mice injected with cell lines derived from E2a:PBX117 or Eμ-RET18 transgenic mice, respectively. All animal studies were approved by the National Cancer Institute Intramural Animal Care and Use Committee.
Human samples
Samples containing ≥90% blasts were obtained from the Austrian study center upon institutional review approval and with approval of the ethics committee. Informed consent for tissue banking and research studies was obtained from patients, their parents, and/or their legal guardians in accordance with the Declaration of Helsinki.
Statistics
Continuous variables were compared using an unpaired 2-tailed Student t test statistic. A 2×2 contingency table and χ2 test with Yates’ correction was used to analyze VH region utilization.
Results
NP23 mice develop a B220− B-lineage ALL
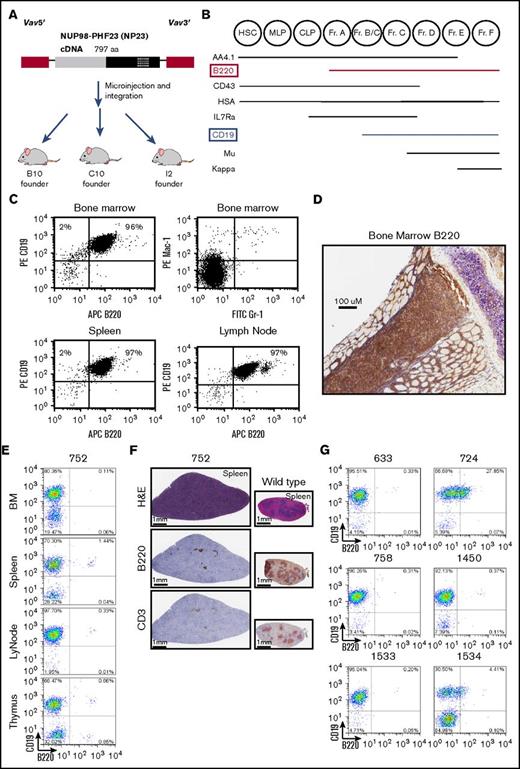
We recently reported that NUP98-PHF23 (NP23) transgenic mice develop acute leukemia with almost complete penetrance by 14 months of age; a diagram of the transgene, which uses Vav1 regulatory elements19 to direct transgene expression to the hematopoietic compartment, is provided in Figure 1A.12 In the initial cohort of mice studied, we detected leukemia in 40 progeny of the B10 founder and 37 progeny of the C10 founder, as well as in 6 additional founder mice.12 Although most mice developed myeloid or T-cell ALL, 1 mouse (founder I2) developed a BCP-ALL with the immunophenotype of a typical BCP-ALL: CD19+B220+sIgm− (Figure 1B-C). Flow cytometry showed a complete lack of normal Mac1+/Gr1+ myeloid cells in the bone marrow and complete replacement with CD19+B220+ B cells; spleen, lymph node, and thymus also showed invasion of CD19+B220+ B cells (Figure 1C). These findings were confirmed by immunohistochemistry showing complete effacement of the bone marrow with B220+ cells (Figure 1D). Comparison with a standard scheme for B-cell differentiation (Figure 1B)20 showed that these cells had matured to at least the fraction BC stage of differentiation, because they had acquired CD19. The diagnosis of B-lineage ALL was further supported by the presence of clonal Igh gene rearrangements (supplemental Figure 1).
A subset of NP23 mice develop a CD19+B220−leukemia. (A) Schematic of the NP23 transgene. Vav 5′ and 3′ regulatory elements are indicated in red, the NUP98 portion of the transgene is gray, and the PHF23 portion is black, with the PHD domain indicated with crosshatches. (B) Schematic of murine bone marrow (BM) B-lymphopoietic differentiation stages, adapted from Hardy and Hayakawa.20 (C) Flow cytometry profile of I2 mouse BM, spleen, and lymph node, stained with the indicated antibodies. (D) Immunohistochemical staining of I2 BM with B220 antibody. (E) Representative flow cytometry profiles of BM, spleen, lymph node (LyNode), and thymus from NP-23 B-lineage ALL 752 using CD19 and B220 antibodies. (F) Hematoxylin and eosin (H&E), B220, and CD3 immunohistochemistry of infiltrated 752 spleen compared with a wild-type (WT) control. (G) B220/CD19 cell populations in additional pro–B-1 ALL samples. Analysis shown is of tissues with the highest purity of malignant cells: 633 BM, 724 BM, 758 BM, 1450 BM, 1533 LyNode, and 1534 BM. CLP, common lymphoid progenitor; Fr, fraction; HSC, hematopoietic stem cell; MLP, multilineage progenitor.
A subset of NP23 mice develop a CD19+B220−leukemia. (A) Schematic of the NP23 transgene. Vav 5′ and 3′ regulatory elements are indicated in red, the NUP98 portion of the transgene is gray, and the PHF23 portion is black, with the PHD domain indicated with crosshatches. (B) Schematic of murine bone marrow (BM) B-lymphopoietic differentiation stages, adapted from Hardy and Hayakawa.20 (C) Flow cytometry profile of I2 mouse BM, spleen, and lymph node, stained with the indicated antibodies. (D) Immunohistochemical staining of I2 BM with B220 antibody. (E) Representative flow cytometry profiles of BM, spleen, lymph node (LyNode), and thymus from NP-23 B-lineage ALL 752 using CD19 and B220 antibodies. (F) Hematoxylin and eosin (H&E), B220, and CD3 immunohistochemistry of infiltrated 752 spleen compared with a wild-type (WT) control. (G) B220/CD19 cell populations in additional pro–B-1 ALL samples. Analysis shown is of tissues with the highest purity of malignant cells: 633 BM, 724 BM, 758 BM, 1450 BM, 1533 LyNode, and 1534 BM. CLP, common lymphoid progenitor; Fr, fraction; HSC, hematopoietic stem cell; MLP, multilineage progenitor.
However, several mice subsequently developed B-lineage leukemia that could not be classified using this standard scheme for B-cell differentiation.20 An example of 1 such animal, #752, is shown in Figure 1E-F. Flow cytometry (supplemental Figure 2 provides an example of gating strategy) of bone marrow, lymph node, spleen, and thymus showed marked invasion of a uniform CD19+/B220− population; close inspection of the flow cytometry profile (Figure 1E) revealed a separate CD19+/B220+ population (presumably from residual nonleukemic B cells), indicating that there was not a technical problem with the B220 antibody.
Immunohistochemical analyses supported the flow cytometry findings, revealing invasion and destruction of the normal splenic architecture by cells that were B220− and CD3− (Figure 1F). Follicle remnant staining with B220 and CD3 can be seen. The CD19+B220− immunophenotype was puzzling; because standard models of murine B-cell differentiation show that B220 expression precedes CD19 expression (Figure 1B),20 all CD19+ cells should also be B220+. This phenomenon was not an oddity present in a single sample; 6 additional mice from the original cohort displayed this CD19+/B220− phenotype (Figure 1G; Table 1). In sum, a total of 7 B-lineage ALLs with a CD19+/B220− phenotype were identified in our initial cohort, a frequency of 6 of 40 and 1 of 37 for the B10 and C10 NP23 founder lines, respectively (Table 1). Coincidentally, 2 of the 7 cases (#758 and #1534) had a concurrent precursor T-cell (pre-T) lymphoblastic lymphoma/leukemia (LBL) characterized by a distinct CD4+/CD8+ population in addition to the B-lineage ALL; we previously observed a similar phenomenon in NP23 mice, which had concurrent AML and pre-T LBL.12
NP23 B-lineage ALL characteristics
| ID . | Age, mo . | Sex . | Malignant immunophenotype . | Infiltrated tissues . | Peripheral blood CBC . | |||
|---|---|---|---|---|---|---|---|---|
| WBC, ×109/L . | HgB, g/dL . | MCV, fL . | PLT, ×109/L . | |||||
| 633 | 8.5 | F | B220−/CD19+/AA4.1+ | BM, Spl, Liv, Kid | 9.7 | 3.3 | 59.1 | 225 |
| 724 | 7.75 | F | B220lo/−/CD19+/AA4.1+ | BM, Spl, LyNo, Thy, Liv, Kid | 102.9 | 4.8 | 67.3 | 202 |
| 752 | 5.75 | M | B220−/CD19+/AA4.1+ | BM, Spl, LyNo, Thy, Liv, Kid | 135.7 | 10.5 | 53.8 | 714 |
| 758 | 10 | F | B220−/CD19+/AA4.1+ | BM, Spl, LyNo | 78.9 | 6.9 | 64.8 | 153 |
| CD4+/CD8+* | Thy | |||||||
| 1450† | 9 | F | B220−/CD19+ | BM, Spl, LyNo | 34.84 | 7.1 | 87.5 | 92 |
| 1533 | 7 | F | B220−/CD19+/AA4.1+ | BM, Spl, LyNo, Liv | nt | |||
| 1534 | 7.5 | F | B220lo/−/CD19+/AA4.1 | BM, Spl, LyNo, Thy, Liv, Kid | nt | |||
| CD4+/CD8+* | BM, Spl, LyNo, Thy, Liv, Kid | |||||||
| ID . | Age, mo . | Sex . | Malignant immunophenotype . | Infiltrated tissues . | Peripheral blood CBC . | |||
|---|---|---|---|---|---|---|---|---|
| WBC, ×109/L . | HgB, g/dL . | MCV, fL . | PLT, ×109/L . | |||||
| 633 | 8.5 | F | B220−/CD19+/AA4.1+ | BM, Spl, Liv, Kid | 9.7 | 3.3 | 59.1 | 225 |
| 724 | 7.75 | F | B220lo/−/CD19+/AA4.1+ | BM, Spl, LyNo, Thy, Liv, Kid | 102.9 | 4.8 | 67.3 | 202 |
| 752 | 5.75 | M | B220−/CD19+/AA4.1+ | BM, Spl, LyNo, Thy, Liv, Kid | 135.7 | 10.5 | 53.8 | 714 |
| 758 | 10 | F | B220−/CD19+/AA4.1+ | BM, Spl, LyNo | 78.9 | 6.9 | 64.8 | 153 |
| CD4+/CD8+* | Thy | |||||||
| 1450† | 9 | F | B220−/CD19+ | BM, Spl, LyNo | 34.84 | 7.1 | 87.5 | 92 |
| 1533 | 7 | F | B220−/CD19+/AA4.1+ | BM, Spl, LyNo, Liv | nt | |||
| 1534 | 7.5 | F | B220lo/−/CD19+/AA4.1 | BM, Spl, LyNo, Thy, Liv, Kid | nt | |||
| CD4+/CD8+* | BM, Spl, LyNo, Thy, Liv, Kid | |||||||
BM, bone marrow; CBC, complete blood count; F, female; HgB, hemoglobin; Kid, kidney; Liv, liver; LyNo, lymph node; M, male; MCV, mean corpuscular volume; nt, not tested; Spl, spleen; Thy, thymus; WBC, white blood cell.
These 2 mice had concurrent pre-T LBL.
C10 line.
Given that the B220 antibody reacts with a specific glycosylated isoform of CD45 (CD45RABC),21 we looked for evidence of this isoform. In all but 1 of the B220− samples, the B220 splice species was either absent or markedly reduced (supplemental Figure 3), supporting the observation that these samples do not stain for the B220 isoform of CD45. Of note, the sample that did display abundant levels of the CD45 RABC messenger RNA (#724) showed a heterogeneous staining with B220 (Figure 1F), leading us to consider these B220− B-lineage ALLs could be classified as B220lo/−. Overall, we were still left with the puzzling observation that these leukemias were predominantly B220−/CD19+ and thus did not follow the standard differentiation scheme for murine B-cell differentiation (Figure 1B).
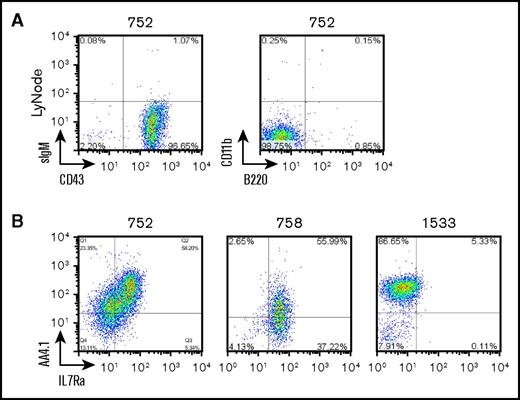
A pro–B-1 B lymphocyte has been identified with the immunophenotype B220lo/−CD19+AA4.1+CD43+,4 leading us to speculate that the B-lineage leukemias we were studying might be pro–B-1 leukemias. Additional staining demonstrated that the malignant cells were negative for common lineage markers including Gr1, CD3, CD4, CD8, Ter119, IgM, and CD11b (Mac1); positive for AA4.1 and CD43; and heterogeneous for Il7rα (Figure 2A-B; Table 1). Il7rα pairs with Crlf2 to form the receptor for thymic stromal lymphopoietin (Tslp), a cytokine that stimulates proliferation of pro–B-1 cells (reviewed by Montecino-Rodriguez and Dorshkind2 ). Consistent with the possibility that these leukemias were of pro–B-1 origin, we found these leukemias expressed Crlf2 using real-time quantitative polymerase chain reaction (PCR) and flow cytometry assays (supplemental Figure 4).
NP23 B-lineage ALLs display features consistent with pro–B-1 cells. (A) NP23 ALL sample 752 stained with indicated antibodies. (B) Samples 752, 758, and 1533 stained with AA4.1 and IL7Ra. LyNode, lymph node.
NP23 B-lineage ALLs display features consistent with pro–B-1 cells. (A) NP23 ALL sample 752 stained with indicated antibodies. (B) Samples 752, 758, and 1533 stained with AA4.1 and IL7Ra. LyNode, lymph node.
NP23 B-lineage ALLs show gene expression signatures similar to those of fetal liver pro B cells
We used gene expression profiles to better characterize these leukemias. Analysis of gene expression arrays from the NP23 B-lineage ALL identified 946 probe sets that were >2-fold overexpressed in NP23 B-lineage ALL compared with WT spleen (GSE54787; supplemental Table 1). Comparison of this gene set with a reference set of gene expression profiles derived from a wide spectrum of purified murine B-cell populations (ImmGen) revealed 4 discrete groups: mature B cells, germinal center B cells, common or mixed lymphoid progenitors, and committed pro– or pre–B cells (supplemental Figure 5A). To confirm the expression array data, we used RNA sequencing (RNA-seq) to identify a set of 646 genes that were >2-fold overexpressed in NP23 B-lineage leukemia compared with WT pro– and pre–B cells (fetal liver CD19+ B220+ cells; supplemental Table 2). Comparison of this data set with the ImmGen expression arrays again revealed the same 4 groups (supplemental Figure 5B). A total of 106 genes were >2-fold overexpressed in both the expression array and RNA-seq comparisons; these are designated NP23 leukemia core genes (supplemental Table 3).
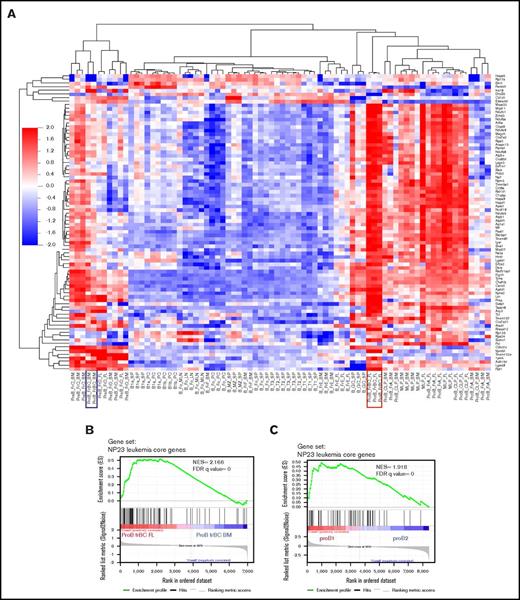
Hierarchical clustering of the NP23 leukemia core genes revealed that the population most similar to the NP23 B-lineage leukemias were fetal liver fraction BC pro B cells; these cells have previously been shown to be precursors for B-1 lymphocytes22 (Figure 3A). Gene set enrichment analysis (GSEA) demonstrated enrichment of the NP23 B-lineage leukemia gene signature in fetal liver fraction BC compared with bone marrow fraction BC (Figure 3B); of note, pro–B-1 cells are known to be enriched in fetal liver compared with bone marrow.22 We next compared the NP23 leukemia core genes identified by combined expression array and RNA-seq with independent, recently described expression profiles of purified pro–B-1 and pro–B-2 cells23 ; GSEA analysis showed that the NP23 B-lineage leukemia gene signature was enriched in pro–B-1 compared with pro–B-2 cells (Figure 3C).
NP23 B-lineage leukemia gene signature is enriched in fetal liver fraction BC and pro–B-1 cells. (A) Heat map shows clustering of 106 NP23 leukemia core genes (>2-fold upregulated in NP23 B-lineage leukemias) in expression array data from isolated B-cell populations (GSE15907). Fetal liver (FL) fraction (Fr) BC profile is boxed in red; bone marrow (BM) fraction BC profile is boxed in blue. (B) GSEA plot showing enrichment of NP23 leukemia core upregulated genes in FL Fr BC compared with BM Fr BC (GSE15907). (C) Enrichment of NP23 leukemia core upregulated genes in pro–B-1 vs pro–B-2 cells23 (GSE81411). FDR, false-discovery rate; LN, lymph node; MLN, mesenteric LN; NES, normalized enrichment score; PC, peritoneal cavity; SP, spleen.
NP23 B-lineage leukemia gene signature is enriched in fetal liver fraction BC and pro–B-1 cells. (A) Heat map shows clustering of 106 NP23 leukemia core genes (>2-fold upregulated in NP23 B-lineage leukemias) in expression array data from isolated B-cell populations (GSE15907). Fetal liver (FL) fraction (Fr) BC profile is boxed in red; bone marrow (BM) fraction BC profile is boxed in blue. (B) GSEA plot showing enrichment of NP23 leukemia core upregulated genes in FL Fr BC compared with BM Fr BC (GSE15907). (C) Enrichment of NP23 leukemia core upregulated genes in pro–B-1 vs pro–B-2 cells23 (GSE81411). FDR, false-discovery rate; LN, lymph node; MLN, mesenteric LN; NES, normalized enrichment score; PC, peritoneal cavity; SP, spleen.
NP23 B-lineage ALLs preferentially use 3′ VH genes
All of the pro–B-1 ALL samples showed clonal Igh gene rearrangements by southern blot analysis or PCR (supplemental Figures 1 and 6). We investigated the use of Igh VH, DH, and JH gene families in these samples by sequencing PCR-amplified VDJ rearrangements. VH gene sequences were identified using GenBank accession number BN000872.1 as a reference24 and D segments as reported by Ye25 (supplemental Table 4). Samples 758 and 1534 had 1 clonal DJ rearrangement and 2 clonal VDJ rearrangements, indicating that there was subclonal heterogeneity within the tumor cell populations. Mouse 758 had 2 clonal VDJ rearrangements that showed an identical DJ junction but distinct VD junctions, suggesting clonal evolution after transformation and expansion of a cell that had undergone a DJ rearrangement. The VH regions used showed a clear bias toward use of 3′ VH gene families; a similar bias has previously been seen in fetal liver B-1 cells2,26 (supplemental Figure 6).
We conclude that these B220lo/−/CD19+ leukemias originated from an immature B cell similar to the BCP LBL defined in the Bethesda proposals.27 However, given that 1) the Lin−/B220lo/−/CD19+/AA4.1+ immunophenotype matches that of a progenitor B-1 cell, 2) the gene expression profile is more similar to that of pro–B-1 cells than any other hematopoietic cell type, and 3) the leukemic clones used 3′ VH regions, we suggest that these leukemias be distinguished from previously defined BCP LBLs and given the designation pro–B-1 ALL.
Spontaneous Bcor mutations occur in pro–B-1 ALL
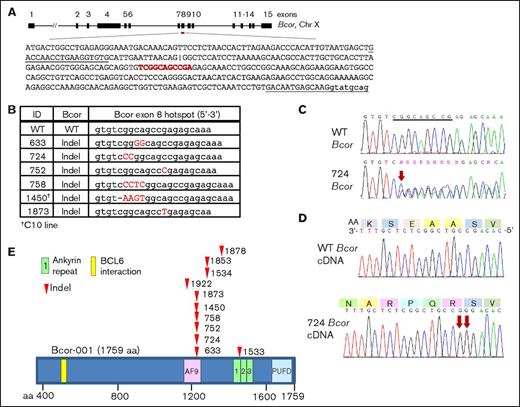
We suspected that acquisition of spontaneous, collaborative mutations was required for malignant transformation of the NP23 pro–B-1s and used whole-exome sequencing (WES) to screen for mutations in 11 NP23 pro–B-1 ALL samples (the 7 described in Table 1 from the initial cohort, plus 4 identified subsequently). Remarkably, WES revealed that all 11 samples had acquired indel (insertion/deletion) mutations leading to the introduction of premature stop codons and truncation of the BCL6 interacting corepressor (Bcor) protein (Figure 4A-B; supplemental Table 5). Of note, 6 of 11 mutations occurred within an 11-bp palindromic hotspot within exon 8 (Figure 4A-B), suggesting an unusual sequence that may be prone to DNA double-strand breaks. Mutations were validated by Sanger sequencing PCR products (Figure 4C). Bcor is located on chromosome X; however, only the mutant Bcor allele was expressed in all 5 pro–B-1 ALLs examined (Figure 4D).
Bcor mutations in NP23 pro–B-1 ALL. (A) Schematic of mouse Bcor transcript (Bcor-001; ENSMUST00000115513), exons 1 to 15. Black upper-case text is exon 8 coding sequence. Underlined sequence indicates PCR primers. An internal alternative exon 8 splice acceptor site is indicated (|). The Bcor 11-bp palindromic mutational hotspot is shown in red. (B) Bcor exon 8 mutations at the exon 8 hotspot; indels shown in red capital letters. (C) WT and 724 pro–B-1 ALL sequence across the mutational hotspot (underlined). Red arrow indicates site of CC dinucleotide insertion. (D) 724 messenger RNA expression (lower panel). Red arrows indicate CC dinucleotide insertion. Predicted amino acid sequence is indicated (upper panel). (E) Schematic of the mouse Bcor protein and mutation sites (arrowheads).
Bcor mutations in NP23 pro–B-1 ALL. (A) Schematic of mouse Bcor transcript (Bcor-001; ENSMUST00000115513), exons 1 to 15. Black upper-case text is exon 8 coding sequence. Underlined sequence indicates PCR primers. An internal alternative exon 8 splice acceptor site is indicated (|). The Bcor 11-bp palindromic mutational hotspot is shown in red. (B) Bcor exon 8 mutations at the exon 8 hotspot; indels shown in red capital letters. (C) WT and 724 pro–B-1 ALL sequence across the mutational hotspot (underlined). Red arrow indicates site of CC dinucleotide insertion. (D) 724 messenger RNA expression (lower panel). Red arrows indicate CC dinucleotide insertion. Predicted amino acid sequence is indicated (upper panel). (E) Schematic of the mouse Bcor protein and mutation sites (arrowheads).
The premature stop codons encode a truncated Bcor protein that retains the BCL6 interaction domain and a portion of the AF9 binding domain but lacks the ankyrin repeats and the PCGF Ub-like fold discriminator (PUFD) domain (Figure 4E). We were unable to detect a truncated Bcor protein in the mutant samples, despite using 2 independent anti-Bcor antibodies (ab135801 and BCoR 186) that recognized epitopes upstream of the mutation hotspot (supplemental Figure 7), suggesting that the mutant protein may be unstable. Indeed, even when artificially overexpressed in 293T cells, only a faint band representing the truncated protein could be detected (supplemental Figure 7). However, it also may be that Bcor expression is below the limit of detection of the antibodies in B cells, because we did not detect Bcor protein in WT spleen either. We also examined 16 NP23 AML samples for Bcor mutations using WES; in contrast to the high frequency of Bcor mutations in the pro–B-1 leukemias, all 16 AML samples were germline for Bcor.28 In addition, WES of the single pre–B-2 ALL sample from the NP23 colony (founder I2) revealed no Bcor mutation.
Jak/Stat pathway activation is associated with pro–B-1 ALL
In addition to acquired Bcor mutations, spontaneous mutations of Jak1, Jak2, or Jak3 were identified by WES and validated by Sanger sequencing in 7 of the 11 samples (supplemental Table 5). WES variant allele frequency suggested a homozygous Jak1 mutation in sample 724 and heterozygous mutations in the others. Sample 633 had a Jak2 point mutation with a variant allele frequency of 14%, suggesting that this was a subclonal progression event. Three of the Jak mutations resided within the kinase catalytic domain, whereas the other 5 lay within the pseudokinase domain, a regulatory region that directly binds the catalytic domain and controls its activation.29 The V657F mutation is similar to the V658F mutation in the pseudokinase domain of JAK1 identified in patients with high-risk BCP-ALL or Down syndrome BCP-ALL,16,30-32 and the Y651H mutation is similar to the Y652H mutation found in patients with T-cell ALL.33,34 The JAK V658F mutation has been reported to result in constitutive activation of STAT3 and STAT5,35 suggesting the involvement of the JAK/STAT signaling pathway (reviewed by Vainchenker and Constantinescu36 ) in the etiology of these pro–B-1 ALLs. Moreover, 1 of the 4 samples that lacked a Jak mutation (#1533) had acquired a Stat5a missense mutation. Phosphorylation of Stat5 was present, and phosphorylated Stat3 (pStat3) was moderately increased as well (supplemental Figure 8). Of note, sample 1533, which showed the most dramatic Stat5 phosphorylation, did not have a Jak mutation but did have an acquired Stat5a mutation (supplemental Table 3).
Patients with BCP-ALL and CRLF2 overexpression have a gene expression profile similar to that of NP23 pro–B-1 ALL and preferentially use B-1 lymphocyte VH segments
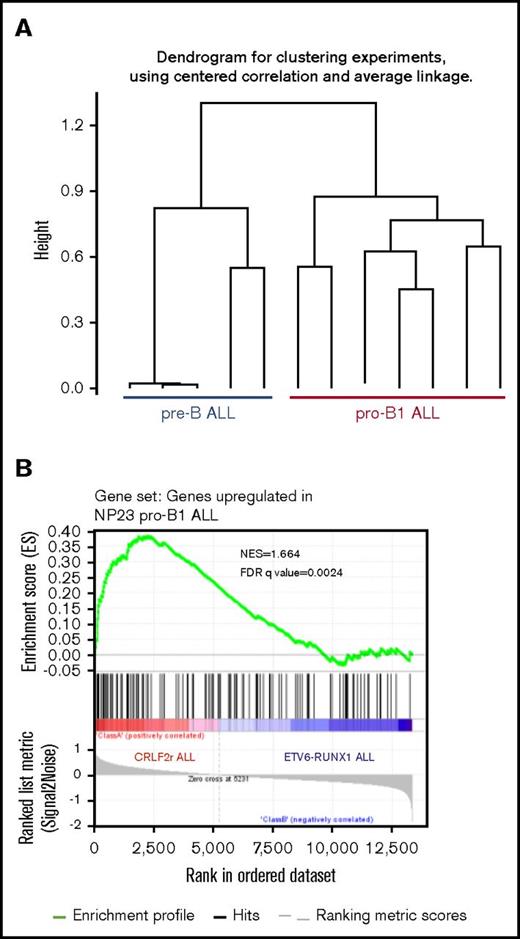
Of note, JAK1/2 mutations in patients with BCP-ALL often occur in the context of aberrant CRLF2 expression,37,38 similar to the Jak1/2 mutations and Crlf2 expression that we have observed in the murine pro–B-1 ALLs. The association of JAK1/2 mutation and CRLF2 activation, along with the observation that murine pro–B-1s are dependent on the Crlf2 ligand (Tslp),4 leads to the intriguing hypothesis that some patients with BCP-ALL may have an ALL of B-1, as opposed to B-2, origin. We used RNA-seq to compare the gene expression profile of 2 murine pre–B-2 ALL models (CD19+B220+CD22+CD43+SIgM−), generated by expression of E2A-PBX117,39 or Eu-RET transgenes,18 with the NP23 pro–B-1 ALL model. Hierarchical clustering clearly separated the murine pre–B-2 ALL and the NP23 pro–B-1 ALL samples (Figure 5A). We identified a gene set that was at least 2-fold overexpressed in pro–B-1 ALL compared with pre–B-2 ALL and used this gene signature in a GSEA comparison of human BCP-ALLs with CRLF2 rearrangements (CRLF2r) with those without CRLF2 rearrangements40 (RUNX1-ETV6 rearrangements and CRLF2 rearrangements are mutually exclusive). As shown in Figure 5B, the murine pro–B-1 upregulated signature was strongly enriched in the CRLF2r group of patients.
NP23 pro–B-1 ALL gene signature is enriched in pediatric ALL with CRLF2 rearrangements. (A) Hierarchical clustering of RNA-seq samples showing that NP23 pro–B-1 ALL expression profile is distinct from that of murine pre–B-2 ALL (taken from Eμ-RET and E2A-PBX mice). (B) GSEA plot showing enrichment of NP23 pro–B-1 ALL gene signature in pediatric ALL carrying CRLF2 rearrangements (GSE26281). The gene signature was generated by comparing NP23 pro–B-1 ALL with mouse pre–B-2 ALL (FPKM FC>2).
NP23 pro–B-1 ALL gene signature is enriched in pediatric ALL with CRLF2 rearrangements. (A) Hierarchical clustering of RNA-seq samples showing that NP23 pro–B-1 ALL expression profile is distinct from that of murine pre–B-2 ALL (taken from Eμ-RET and E2A-PBX mice). (B) GSEA plot showing enrichment of NP23 pro–B-1 ALL gene signature in pediatric ALL carrying CRLF2 rearrangements (GSE26281). The gene signature was generated by comparing NP23 pro–B-1 ALL with mouse pre–B-2 ALL (FPKM FC>2).
Pro–B-1 lymphocytes have not yet been identified in humans, making it difficult to compare the immunophenotype of normal human pro–B-1 lymphocytes with that of patients with BCP-ALL and CRLF2 rearrangement. However, mature human B-1 lymphocytes have a restricted VH region usage.10 We reasoned that if patients with BCP-ALL and CRLF2 activation showed the same preference for VH regions as did the mature human B-1 lymphocytes, that observation would further support the hypothesis that a BCP-ALL with CRLF2 activation is derived from pro–B-1 lymphocytes. We sequenced clonal IGH gene rearrangements from 23 patients with BCP-ALL and overexpression of CRLF2 and from 28 patients from a genetically distinct group of BCP-ALLs (ETV6-RUNX1) that did not show CRLF2 overexpression. Table 2 shows that 17 of the 23 patients with CRLF2 overexpression used 1 of the B-1–preferred VH gene regions (highlighted in red), whereas only 10 of 28 patients without CRLF2 overexpression used 1 of these preferred V regions (2-tailed χ2P = .0065). In addition, VH3-30, which was the most commonly used VH region in mature human B-1 cells, accounting for >20% of the total,10 was also the single most commonly used VH region in patients with BCP-ALL and CRLF2 overexpression, being used in 4 (17%) of 23 patients. Furthermore, VH3-30 and VH3-74, the 2 most commonly used VH regions in patients with CRLF2-overexpressing BCP-ALL (Table 2), are markedly overrepresented in the immune response to pneumococcal polysaccharides,41 a response reported to be mediated by B-1 lymphocytes.
VH gene use in 2 genetic subgroups of patients with BCP-ALL
| Patient no. . | IGH V region . | Preferred B-1 V region? . |
|---|---|---|
| CRLF2 overexpression | ||
| 360 | IGHV3-74 | + |
| 365 | IGHV3-23, IGHV3-49 | + |
| 379 | IGHV3-74 | + |
| 400 | IGHV3-33, IGHV3-30 | + |
| 552 | IGHV1-3, IGHV3-11, IGHV3-33 | |
| 569 | IGHV4-39 | + |
| 715 | IGHV2-70 | |
| 802 | IGHV4-34, IGHV3-30 | + |
| 833 | IGHV4-39 | + |
| 839 | IGHV3-71 | |
| 841 | IGHV3-41, IGHV2-5 | |
| 873 | IGHV4-61, IGHV3-66, IGHV1-69 | + |
| 887 | IGHV3-74, IGHV2-70, IGHV1-69 | + |
| 903 | IGHV1-8 | |
| 948 | IGHV3-30 | + |
| 957 | IGHV1-46, IGHV3-13, IGHV4-30 | + |
| 961 | IGHV3-74, IGHV3-13 | + |
| 1047 | IGHV4-39, IGHV3-7, IGHV2-26 | + |
| 1060 | IGHV4-59 | + |
| 1063 | IGHV3-71, IGHV3-30 | + |
| 1089 | IGHV3-22, IGHV3-13 | |
| 1097 | IGHV2-26 | + |
| 1101 | IGHV3-9 | + |
| ETV6-RUNX1 fusion | ||
| 960 | IGHV3-21 | |
| 968 | IGHV3-13 | |
| 985 | IGHV3-23, IGHV2-5 | + |
| 991 | IGHV6-1 | |
| 997 | IGHV2-5, IGHV4-61 | |
| 999 | IGHV3-22 | |
| 1000 | IGHV3-15 | |
| 1029 | IGHV5-3 | |
| 1034 | IGHV1-2, IGHV1-2 | + |
| 1042 | IGHV4-61, IGHV2-5, IGHV3-41 | |
| 1052 | IGHV3-7 | |
| 1053 | IGHV7-4,IGHV2-5, IGHV3-30 | + |
| 1054 | IGHV1-2, IGHV2-5 | + |
| 1059 | IGHV3-74 | + |
| 1061 | IGHV3-53, IGHV1-69 | + |
| 1075 | IGHV3-74 | + |
| 1081 | IGHV3-13 | |
| 1084 | IGHV1-8 | |
| 1115 | IGHV1-69, IGHV4-55 | + |
| 1118 | IGHV3-71, IGHV2-5 | |
| 1120 | IGHV3-11, IGHV4-59 | + |
| 1122 | IGHV2-70, IGHV3-23 | + |
| 1133 | IGHV3-33 | |
| 1139 | IGHV3-71, IGHV1-45 | |
| 1147 | IGHV3-13 | |
| 1153 | IGHV3-7 | |
| 1162 | IGHV2-5 | |
| 1170 | IGHV6-1 | |
| Patient no. . | IGH V region . | Preferred B-1 V region? . |
|---|---|---|
| CRLF2 overexpression | ||
| 360 | IGHV3-74 | + |
| 365 | IGHV3-23, IGHV3-49 | + |
| 379 | IGHV3-74 | + |
| 400 | IGHV3-33, IGHV3-30 | + |
| 552 | IGHV1-3, IGHV3-11, IGHV3-33 | |
| 569 | IGHV4-39 | + |
| 715 | IGHV2-70 | |
| 802 | IGHV4-34, IGHV3-30 | + |
| 833 | IGHV4-39 | + |
| 839 | IGHV3-71 | |
| 841 | IGHV3-41, IGHV2-5 | |
| 873 | IGHV4-61, IGHV3-66, IGHV1-69 | + |
| 887 | IGHV3-74, IGHV2-70, IGHV1-69 | + |
| 903 | IGHV1-8 | |
| 948 | IGHV3-30 | + |
| 957 | IGHV1-46, IGHV3-13, IGHV4-30 | + |
| 961 | IGHV3-74, IGHV3-13 | + |
| 1047 | IGHV4-39, IGHV3-7, IGHV2-26 | + |
| 1060 | IGHV4-59 | + |
| 1063 | IGHV3-71, IGHV3-30 | + |
| 1089 | IGHV3-22, IGHV3-13 | |
| 1097 | IGHV2-26 | + |
| 1101 | IGHV3-9 | + |
| ETV6-RUNX1 fusion | ||
| 960 | IGHV3-21 | |
| 968 | IGHV3-13 | |
| 985 | IGHV3-23, IGHV2-5 | + |
| 991 | IGHV6-1 | |
| 997 | IGHV2-5, IGHV4-61 | |
| 999 | IGHV3-22 | |
| 1000 | IGHV3-15 | |
| 1029 | IGHV5-3 | |
| 1034 | IGHV1-2, IGHV1-2 | + |
| 1042 | IGHV4-61, IGHV2-5, IGHV3-41 | |
| 1052 | IGHV3-7 | |
| 1053 | IGHV7-4,IGHV2-5, IGHV3-30 | + |
| 1054 | IGHV1-2, IGHV2-5 | + |
| 1059 | IGHV3-74 | + |
| 1061 | IGHV3-53, IGHV1-69 | + |
| 1075 | IGHV3-74 | + |
| 1081 | IGHV3-13 | |
| 1084 | IGHV1-8 | |
| 1115 | IGHV1-69, IGHV4-55 | + |
| 1118 | IGHV3-71, IGHV2-5 | |
| 1120 | IGHV3-11, IGHV4-59 | + |
| 1122 | IGHV2-70, IGHV3-23 | + |
| 1133 | IGHV3-33 | |
| 1139 | IGHV3-71, IGHV1-45 | |
| 1147 | IGHV3-13 | |
| 1153 | IGHV3-7 | |
| 1162 | IGHV2-5 | |
| 1170 | IGHV6-1 | |
Discussion
We have characterized a subset of NP23 transgenic mice that developed an aggressive, invasive leukemia with an immunophenoytpe (Lin−/B220−/CD19+/AA4.1+) that closely matched that of a pro–B-1 cell. Although similar to the BCP LBL described in the Bethesda proposals27 for classifying murine lymphoid malignancy, these cells were B220− and thus were inconsistent with any known B-2 pro– or pre–B cell.20 To the best of our knowledge, leukemias of pro–B-1 cells have not previously been described in mice or humans, and we designate these as pro–B-1 ALLs.
Gene expression profiling further supported the assertion that the NP23 B-lineage ALL arose from pro–B-1 cells. We used both gene expression arrays as well as RNA-seq to identify genes that were consistently upregulated in NP23 B-lineage ALL compared with WT B cells. Comparison of the genes most highly expressed in the pro–B-1 ALLs with gene expression profiles of murine B cells revealed that the NP23 pro–B-1 ALL was most similar to fetal liver pro-B fraction BC, more so than adult bone marrow pro-B fraction BC. Given that fetal liver fraction BC has been reported to be a source of B-1 B cells,22 these observations again support a pro–B-1 cell as the normal counterpart of the NP23 B-lineage leukemias. Moreover, direct comparison of the genes most highly upregulated in NP23 pro–B-1 ALL with an independent RNA-seq data set revealed that the NP23-upregulated gene profile was more similar to that of pro–B-1 cells than pro–B-2 cells, further supporting the assertion that the NP23 B-lineage ALLs were of pro–B-1 origin.23
Clonality was evident by DJ or VDJ rearrangement in the B cells and demonstrated a bias toward the use of 3′ VH gene families VH7183 and VHQ52, a phenomenon observed in fetal B-1 cells (reviewed by Montecino-Rodriguez and Dorshkind2 and Hardy26 ), further supporting the pro–B-1–cell origin. Using the nomenclature and VH family designation provided by ImMunoGeneTics, we noted that 10 of the 11 VH regions used belonged to the VH1, VH2, or VH5 families; these 3 families made up >70% of VH regions used by B-1a cells in 1 report.42 Furthermore, use of the DH segments DFL16.1, DSP, DST4, and DQ52 is characteristic of fetal B-1a cells42 ; Table 2 shows that all 13 of the clonal rearrangements used 1 of these 4 D segments. It should be noted that B-1 B cells have historically been thought to have little or no N region addition at VDJ junctions20,43 ; however, recent studies have clearly shown that subsets of pro–B-1 cells, specifically B-1a cells, have abundant N region addition.42,44 In 1 study, B-1a cells derived from bone marrow CD19+ cells showed N region addition,42 and a second study demonstrated that the PC1lo subset of B-1a cells had both frequent N region additions and preferential usage of the VH5 family.44 Taken together, characteristics of Igh locus VDJ rearrangement further support the contention that these leukemias represent malignancies of pro–B-1 origin.
Remarkably, all 11 pro–B-1 ALL samples had acquired similar Bcor mutations during the process of transformation. All Bcor mutations were short indels, and all encoded a truncated form of the Bcor protein. Bcor was initially identified as a transcriptional corepressor of Bcl645,46 and interacts with a set of Polycomb group proteins to form a Polycomb repressor 1-like (PRC1-like) complex predicted to exert histone ubiquitylation and demethylation enzymatic functions.47,48 Somatic frameshift and nonsense mutations of BCOR have previously been found to be distributed throughout the BCOR protein in AML,13 bone sarcomas,49 chronic myelomonocytic leukemia and myelodysplastic syndrome,14 and pediatric rhabdomyosarcoma.50 Because BCOR is X linked, and BCOR mutations are often frameshift mutations, it has been suggested that BCOR loss of function may provide a competitive advantage during malignant transformation. This hypothesis is supported by our failure to detect a truncated Bcor protein in the pro–B-1 samples. In addition, a recent report demonstrated an acquired Bcor frameshift mutation in a murine pre–B-2 ALL sample.51 Alternatively, the observation that the Bcor mutations were tightly clustered in the pro–B-1 ALL samples might suggest that there is a specific gain of function conferred by a truncated Bcor protein that retains the Bcl6 interaction domain but lacks the ankyrin repeats and PUFD domain.
Eight of 11 pro–B-1 ALLs had acquired Jak1/2/3 or Stat5a mutations, and all samples examined showed evidence of constitutive Stat signaling, based on increased pStat3 or pStat5. The combination of the NP23 transgene along with spontaneous Bcor and Jak signaling pathway mutations in these pro–B-1 leukemias provides experimental support for a recently published model of high-risk ALL that was based on whole-genome sequencing of patients with BCP-ALL.52 In this model, an initiating event leads to increased self-renewal of pro B cells, followed by a mutation that leads to a block in B-cell differentiation and a mutation that leads to increased proliferation. Experimental support for this model has recently been documented for pre–B-2 ALL but not pre–B-1 ALL.51,53 In the case of NP23 pro–B-1 ALL, the initial event leading to increased self-renewal is the NP23 transgene, introduced into the mouse germline. Although NP23 fusions are rare in B-lineage ALL, the NP23 fusion leads to overexpression of Hoxa cluster genes, the overexpression of which is associated with increased stem-cell self-renewal.12,54 The spontaneous, acquired mutations in Bcor and Jak1/2 lead to a block in B-cell differentiation and hyperproliferation, respectively. This constellation of findings provides experimental support for a hypothesis in which mutations in 3 critical pathways, namely stem-cell self-renewal, impaired B-cell differentiation, and Jak/Stat cytokine signaling, collaborate to induce BCP-ALL.
There are interesting parallels between the pro–B-1 ALL that develops in NP23 transgenic mice and CRLF2-associated BCP-ALL in humans, which represents ∼5% of all childhood BCP-ALLs.37 Both are associated with collaborative mutations in JAK1 or JAK2, both show CRLF2 overexpression (indeed, murine B-1 progenitors have been shown to be dependent on TSLP, which is the ligand for CRLF2), and the gene expression profile of murine NP23 pro–B-1 ALL is markedly more similar to that of CRLF2r ALL than non-CRLF2r ALL. Finally, both murine NP23 pro–B-1 ALL and human CRLF2r ALL show preferential usage of a VH region preferred by B-1 lymphocytes. Taken together, these findings lead to the intriguing hypothesis that patients with BCP ALL and CRLF2 overexpression may have malignancies of pro–B-1 origin. We suggest that the NP23 transgenic mouse will provide an important model for the study of B-1 B-cell development, as well as the molecular and genetic events that lead to pro–B-1 ALL.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Michael Kuehl, Andre Nussenzweig, Sandy Morse, Jack Shern, and current and former members of Aplan laboratory for insightful discussions. The authors also thank the National Cancer Institute (NCI) Sequencing Core for Sanger sequencing, the NCI Transgenic Core for generation of transgenic mice, Shelley Hoover and Mark Simpson of the NCI Molecular Pathology Unit for assistance with slide imaging, and Maria Jorge for excellent animal husbandry and Vivian Bardwell for the kind gift of the anti-Bcor antibody.
This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health (grant numbers ZIA SC 010378 and BC 010983).
Authorship
Contribution: P.D.A., S.M.G., and L.G. conceived and designed the project; S.M.G., L.G., E.J., S.K., M.P., R.L.W., J.D., and S.F. performed experiments; S.M.G., L.G., Y.J.Z., S.B., Y.J.C., T.J.F., R.P.-G., P.S.M., and P.D.A. analyzed the data; all authors reviewed drafts of the manuscript; and P.D.A. wrote the final draft of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter D. Aplan, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Building 37, 37 Convent Dr, Bethesda, MD 20892; e-mail: aplanp@mail.nih.gov.
References
Author notes
*S.M.G. and L.G. contributed equally to this work and should be considered joint first authors.