Key Points
The goal was to compare targeted MRM proteomics with conventional assays to assess concentration levels of coagulation- and fibrinolysis-related proteins.
MRM offers higher sensitivity and multiplicity and good ability to leverage measurements to discriminate groups using unsupervised clustering.
Abstract
The plasma levels of pro- and anticoagulant proteins are important markers for venous thrombosis (VT) risk and can be affected by both genetic and acquired factors, including cancer. Generally, these markers are measured using activity- or antibody-based assays. Targeted proteomics with stable-isotope–labeled internal standards has proven adept at the rapid, multiplex, and precise quantification of proteins in complex biological samples such as plasma. We used liquid chromatography coupled to multiple reaction monitoring (MRM) mass spectrometry to evaluate the concentrations of 31 coagulation- and fibrinolysis-related proteins in plasma from 25 healthy controls, 25 patients with VT, and 25 patients with VT who were also diagnosed with cancer. The concentration level of 1 to 3 proteotypic peptides per protein was determined, and all samples were previously characterized using traditional antibody- or activity-based methods. When comparing the conventional and the MRM strategies, the mean Pearson correlation for the 13 proteins (covered by 36 target peptides) shared between the 2 approaches was 0.77, indicating a good correlation. Additionally, MRM offers higher sensitivity (mean regression slope, 0.81), higher multiplicity in a single run, and good ability to leverage all measurements to discriminate groups using unsupervised clustering, which identified vitamin K antagonist users as well as patients with VT and cancer. The data collected using MRM show that the combination of coagulation factor levels yields signature information on VT and cancer, which was not obvious from a single measurement. These results encourage the further validation and investigation of MRM in profiling protein signature of disease.
Introduction
The plasma levels of functional pro- and anticoagulant proteins, as well as of pro- and antifibrinolytic factors, are important markers for venous thrombosis (VT)1,2 risk and can be affected by both genetic and acquired factors.3-9 Generally, these markers are measured in both clinical chemistry and research laboratories using activity- or antibody-based assays. Although they provide high sensitivity and accuracy, these methods require a relatively large volume of plasma and are costly.
A recent alternative method to measure protein concentration levels is targeted quantitative mass spectrometry–based proteomics, which is increasingly used to quantify the protein contents of biological fluid and tissue samples.10-13 Targeted mass spectrometry–based proteomics enables the protein profiling of biologically complex samples as well as accurate comparisons of the protein content between samples.14-16 A targeted proteomic experiment typically uses a bottom-up workflow based on liquid chromatography–mass spectrometry (LC-MS) analysis of peptides to measure either relative or absolute concentrations of the target proteins in the samples. For the most precise quantification, the absolute concentrations of proteotypic peptides are measured by adding stable-isotope–labeled internal standards to the sample.16,17 An internal standard peptide is chemically synthesized and has identical sequence as the endogenous peptide; however, 1 amino acid has incorporated heavy isotopes of carbon and/or nitrogen (ie, carbon-13 and nitrogen-15), giving it a predetermined mass shift that can be resolved using a mass spectrometer. This allows us to measure the relative abundance of both heavy and light forms and to determine the amount of endogenous peptide by knowing exactly how much heavy-labeled peptide is spiked in.
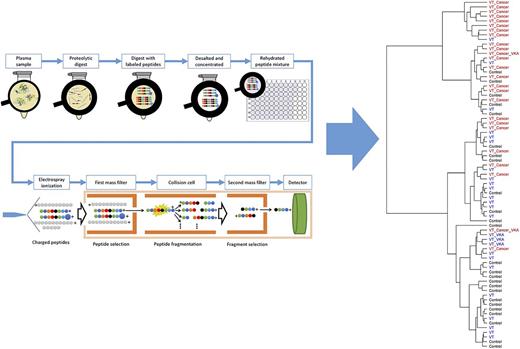
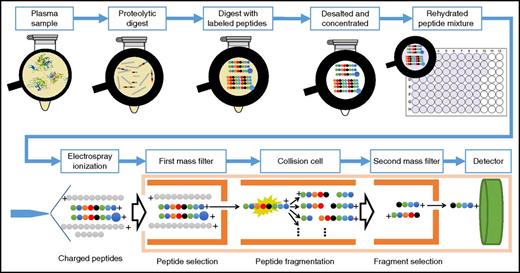
To obtain best sensitivity in the quantification, it is common to use a triple-quadrupole13,18 mass spectrometer and operate it in the targeted-ion mode. Here, the peptide liquid chromatography eluate is measured by multiple reaction monitoring (MRM; also known as selected reaction monitoring). MRM is a type of tandem MS and involves 2 main stages of mass analysis. In these stages, a precursor (ie, a peptide) is selected and fragmented, and its fragment ions are sent to the second mass analyzer, allowing us to record responses of the selected transitions of each of the endogenous as well as heavy-labeled peptides, which finally enables the absolute quantitation of the protein (Figure 1).
A typical LC-MRM/MS experiment. In a standard absolute quantitative experiment, proteins are first extracted from the biological sample and then denatured, reduced, and enzymatically digested (usually with trypsin). The digest is spiked with a known amount of stable-isotope–labeled standard (SIS) peptide corresponding to the targeted peptides from the targeted proteins. The mixture is then submitted for analysis by MS, where the endogenous and isotopically labeled peptides are explicitly identified by retention time and peak shape, precursor and product ion mass-to-charge ratio, and fragment ion ratios. In the mass spectrometer, specific precursor ions are selected in the first mass analyzer according to their mass-to-charge ratio; they are then fragmented by collision-induced dissociation in the collision cell and mass filtered in the second mass analyzer; only the targeted fragment ion reaches the detector. Finally, comparison of the signal from the endogenous peptide and the known amount of isotopically labeled internal standard provides the absolute quantitation.
A typical LC-MRM/MS experiment. In a standard absolute quantitative experiment, proteins are first extracted from the biological sample and then denatured, reduced, and enzymatically digested (usually with trypsin). The digest is spiked with a known amount of stable-isotope–labeled standard (SIS) peptide corresponding to the targeted peptides from the targeted proteins. The mixture is then submitted for analysis by MS, where the endogenous and isotopically labeled peptides are explicitly identified by retention time and peak shape, precursor and product ion mass-to-charge ratio, and fragment ion ratios. In the mass spectrometer, specific precursor ions are selected in the first mass analyzer according to their mass-to-charge ratio; they are then fragmented by collision-induced dissociation in the collision cell and mass filtered in the second mass analyzer; only the targeted fragment ion reaches the detector. Finally, comparison of the signal from the endogenous peptide and the known amount of isotopically labeled internal standard provides the absolute quantitation.
The main advantages of absolute quantification using the LC-MRM/MS approach are that many analytes can be measured in 1 run and that the sample volume/amount required is low when compared with other techniques. Typically, 3 to 5 μL of plasma is required for targeted proteomics. Furthermore, when the sequence of the protein and the background proteome is known, designing an MRM assay is often straightforward, with selection of the proteotypic peptides to function as surrogate for that protein.19 The selection of these peptides follows various rules, including absence of posttranslational modifications, specific lengths for peptides, and the possibility to synthesize heavy-labeled analogous peptides, among others. This allows us to develop quantitative assays for theoretically any protein and also gives us the advantage of knowing exactly which part of the protein we are quantifying, in contrast to antibody assays, where the epitope location is often unknown. In addition, it allows us to quantify a protein with multiple peptides stretching over its sequence, which offers the possibility of protein chain quantitation. Over the last few years, there has been an increase in the multiplicity of MRM/MS peptide panels. Currently, up to 240 peptides with 3 transition ions per peptide form (ie, endogenous and heavy) can be monitored in a single analytical run of 30 to 40 minutes per blood plasma sample.
We here report the results from a multiplexed targeted proteomic LC-MRM/MS evaluation of the concentrations of 31 coagulation- and fibrinolysis-related proteins in the collected plasma of healthy participants and participants with disease.
Materials and methods
In a blinded study, we analyzed plasma samples from 25 healthy controls, 25 patients with VT, and 25 patients with VT who were also diagnosed with cancer. The samples were obtained from a previously well-characterized and analyzed cohort where the concentrations of 15 of the 31 target proteins were evaluated using antibody- (enzyme-linked immunosorbent assay) or activity-based assays.20 Using SIS peptides, we measured the concentration level of up to 3 proteotypic peptides per protein, with a total of 76 peptides representing all 31 proteins. Both the conventional antibody and activity assay measurements as well as the new MRM protein concentration were determined in aliquots from the same plasma samples, allowing direct comparison of the approaches. In the following sections, we include details about the cohort and the methods used.
Patients
A total of 75 samples were obtained from the Multiple Environmental and Genetic Assessment of Risk Factors for Venous Thrombosis (MEGA) case-control study, which has been previously described.20 Briefly, patients age 18 to 70 years, with a first episode of deep venous thrombosis or pulmonary embolism between 1 March 1999 and 31 August 2004 were included (n = 4956). Age- and sex-matched controls were partners of patients (n = 3297) or recruited by random-digit dialing. Randomly, 25 controls, 25 patients with VT, and 25 patients with VT who were also diagnosed with cancer were selected for a targeted proteomic approach. This study was approved by the Medical Ethics Committee of the Leiden University Medical Center, which is a legally recognized committee. All participants provided written informed consent.
Determining plasma protein level using antibody and activity assays
Levels of coagulation factors fibrinogen, II, V, VII, VIII, IX, X, XI, XII, XIII, von Willebrand (VWF), antithrombin, protein C, protein S, plasminogen, tissue plasminogen activator, α-2 antiplasmin, and plasminogen activator inhibitor-1 were measured in plasmas taken at least 3 months after the event.
The detailed analysis was described previously by Blom et al20 ; briefly, blood samples were drawn into vacuum tubes containing 0.1 volume of 0.106 M of trisodium citrate and centrifuged at 2000g for 10 minutes at 4°C, after which plasma was aliquoted, frozen, and stored at −80°C. All assays were performed in automated machines by laboratory technicians who were unaware of the case-control status of the samples. Prothrombin (factor II) activity and factor VII activity were measured with a mechanical clot detection method on a STA-R coagulation analyzer following the instructions of the manufacturer (Diagnostica Stago, Asnieres, France). Levels of factor IX antigen, factor X antigen, factor V antigen, factor XI antigen, and total protein S were determined by enzyme-linked immunosorbent assay. Fibrinogen activity was measured on the STA-R analyzer according to the methods of Clauss. VWF antigen was measured with the immunoturbidimetric method using the STA Liatest kit (rabbit anti-human VWF antibodies), following the instructions of the manufacturer. Measurement of antithrombin and protein C levels was performed with a chromogenic assay on the STA-R analyzer. α-2 antiplasmin and plasminogen activity was measured by using chromogenic assays (STA Stachrom; Diagnostica Stago) performed on a STA-R coagulation analyzer with the use of a commercial calibration standard (Diagnostica Stago).
Determining plasma protein level using LC-MRM/MS assay
Targeted MS-based proteomic assay was designed for 31 proteins covered by 76 surrogate peptides. The proteins and their surrogate peptides are listed in supplemental Table 1. Fifteen of these proteins were evaluated previously by antibody or activity assays (as described in previous paragraph); 16 additional proteins that were not measured by antibody or activity assays were included to demonstrate MRM multiplicity and because of their role in the coagulation process (supplemental Table 2). The peptides for the MRM assays were selected by PeptidePicker19 and synthesized at the University of Victoria Proteomics Centre. The sample preparation was performed in an automated manner using a Tecan Freedom Evo 150 robot. The deoxycholate-based preparation protocol used was developed and applied previously.21 For our study, 30 μL of 10-fold diluted undepleted plasma was added to 174.5 μL of 25 mM ammonium bicarbonate and 30 μL of 10% (weight/volume) sodium deoxycholate for protein denaturation. Disulfide bonds were reduced with a 26.1-μL addition of 50 mM TCEP (tris(2-carboxyethyl)phosphine) for a final concentration of 5 mM. The samples were incubated in dry air at 60°C for 30 minutes, followed by alkylation of free sulfhydryl groups by adding 29 μL of 100 mM iodoacetamide to give a final concentration of 10 mM and incubating at 37°C for 30 minutes in the dark in a dry-air incubator. To prevent alkylation of other residues, the remaining iodoacetamide was quenched by adding 29 μL of 100 mM dithiothreitol and incubated at 37°C in a dry-air incubator for 30 minutes. Proteolytic digestion was then initiated by adding an aliquot of TPCK (L-(tosylamido-2-phenyl) ethyl chloromethyl ketone)–treated bovine trypsin (Worthington, Lakewood, NJ) at a 10:1 ratio of substrate to enzyme. Digestion was allowed to proceed overnight for 16 hours at 37°C. On completion of digestion, the acidified SIS peptide mixture was added. The samples were prepared by combining aliquots of the plasma tryptic digest (117.5 μL) with the SIS peptide mixture (30 μL) and then adding 52.5 μL of 1.9% formic acid (FA) to each standard. After centrifugation at 12 000g for 10 minutes, 133.4 μL of the supernatant was removed from the acid-insoluble sodium deoxycholate and then desalted and concentrated by solid-phase extraction with an Oasis HLB cartridge using traditional vacuum manifold processing. The extractions were performed with vacuum bleed valve set to −25 kPa and a <1 mL/min flow rate. The eluted samples were then frozen, lyophilized to dryness overnight, and rehydrated in 0.1% FA to give a final concentration of 1 μg/μL (assuming an initial plasma protein concentration of 70 mg/mL) before injection. The final SIS peptide concentration for each sample is listed in supplemental Table 1 and was used to calculate the concentration of the analogous endogenous peptide.
The LC/MRM-MS experiments were performed as 1-point measurement estimation22 of each sample by reversed-phase ultrahigh-pressure LC on a 1260 Infinity LC system using a Zorbax Eclipse Plus C18 Rapid Resolution HD column (150 × 2.1 mm; 1.8-μM particles; Agilent Technologies, Palo Alto, CA). The column was maintained at 50°C, and the autosampler was kept at 4°C. Mobile-phase compositions of 0.1% FA in water for solution A and 100% acetonitrile in 0.1% FA for solution B at a flow rate of 0.4 mL/min were used. The gradients were 0:3, 1.5:7, 16:15, 18:15.3, 33:25, 38:45, 39:90, 42.9:90, and 43:3 (time, %B). The LC system was interfaced to a 6490 triple-quadrupole mass spectrometer (both from Agilent Technologies) via a standard-flow electrospray ionization source.
Data analysis
The MRM peak inspection and integration were performed by MassHunter Quantitative Analysis software. The best of the 3 acquired transitions was used as quantifier to calculate the SIS and endogenous responses for single-point measurement.22 For proteins represented with >1 surrogate peptide, the R2 and Pearson correlation coefficient between each 2 peptides for all 75 samples were determined. For all proteins that had concentration levels determined previously by antibody or activity assays, we also calculated correlation with the determined concentration level from the LC-MRM/MS approach. For those proteins, we calculated the R2 and Pearson correlation coefficient (after centralizing and scaling both measurements). Unsupervised cluster analysis was performed on the concentrations determined by LC-MRM/MS. The Ward agglomerative clustering method was used to perform the clustering.23
Results
MRM data inspection and quality
All peptide responses were inspected for interference and correct integration. In all 75 plasma samples, we were able to register interference-free peptide responses from 76 of the 79 measured. Two peptides from factor XI (ie, EDTSFFGVQEIIIHDQYK, VYSGILNQSEIK) and 1 peptide from protein Z (ie, APDLQDLPWQVK) failed for all samples. Of the 75 × 76 measurements (ie, 5700 surrogate peptide abundance evaluations), we were able to determine 5374, whereas 326 did not produce a signal. These 326 were kept in the initial analyses because the study was blinded, and it was not possible to know whether to the lack of signal was a result of condition-related protein abundance or measurement artifact. After lifting the blinding, further inspection of the data showed that there was no evidence that these 326 measurements were related to condition (ie, they were distributed in all 3 sample groups); supplemental Table 3 lists the peptides without signal with 228 of the 326 in 2 proteins: carboxypeptidase B2 and VWF.
To further investigate the quality of the acquired data, for each protein measured by >1 peptide, the pairwise correlation between these peptides was calculated. The peptide-peptide correlations showed a mean Pearson correlation of 0.86. This is considered to indicate that the measured peptide concentration levels were representative of the protein abundance and its variation between samples. In this evaluation, vitamin K–dependent protein Z protease inhibitor and coagulation factor IX showed poor peptide-peptide correlation because of the relatively large number of samples without signal for their peptides ETSNFGFSLLR and VSVSQTSK, respectively (supplemental Table 3).
Correlation between antibody/activity assays and MRM
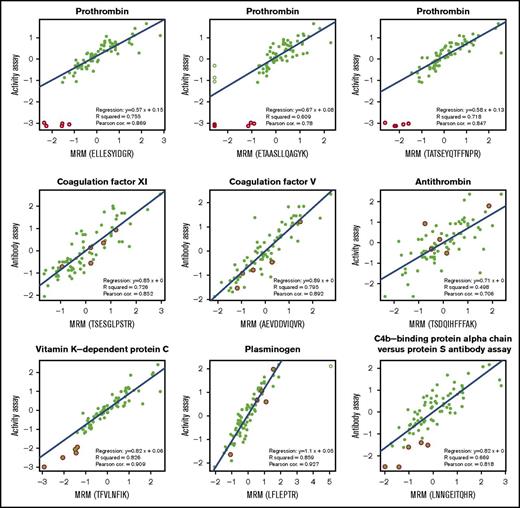
We compared the MRM peptide level responses and available activity or antibody measurements for proteins belonging to the coagulation pathway. For the 13 proteins measured by antibody/activity assay, we measured the levels of 36 surrogate peptides, where fibrinogen is measured by 3 peptides for each protein chain summing up to 9 peptides in total. Figure 2 shows 9 examples of various correlations between MRM and antibody/activity assay. We here present a few examples of proteins with various characteristics, such as being pro- or anticoagulant or profibrinolytic, being an enzyme or cofactor, requiring vitamin K for synthesis or not, and having conventional assay be antibody or activity based.
The correlation between the protein concentrations obtained by MRM and previously obtained activity level for factors II and X. Data are normalized on both axes. Empty symbols are statistical outliers; values originating from patients who used vitamin K antagonists are highlighted in red; the blue line is the regression line. All data points except statistical outliers were considered for the correlations, except for vitamin K–dependent protein C, in which the values in red were also omitted in the calculation of correlation.
The correlation between the protein concentrations obtained by MRM and previously obtained activity level for factors II and X. Data are normalized on both axes. Empty symbols are statistical outliers; values originating from patients who used vitamin K antagonists are highlighted in red; the blue line is the regression line. All data points except statistical outliers were considered for the correlations, except for vitamin K–dependent protein C, in which the values in red were also omitted in the calculation of correlation.
The first example is prothrombin, shown with its 3 measured MRM peptides. All 3 peptides showed high correlations with the activity assay measurements, with Pearson correlation coefficient values of 0.78, 0.85, and 0.87. This was after excluding statistical outliers of ≥ 3 interquartile ranges (Figure 2 open symbols), which were explained after unblinding by the use of vitamin K antagonists by these 5 patients (Figure 2). The same outliers were observed for the other vitamin K–dependent factors VII, IX, and X.
The second example is coagulation factor XI, which unlike prothrombin was measured by an antibody assay instead of activity assay. In MRM, it was measured by 1 peptide (TSESGLPSTR), which showed a Pearson correlation of 0.85 with the antibody assay measurement. The third is coagulation factor V, which was measured by 3 peptides in the MRM method with correlations of 0.8, 0.86, and 0.89 with its antibody assay (1 peptide is shown in Figure 2; all peptides are listed in Table 1).
Target proteins measured by MRM and their corresponding surrogate peptides
| Protein . | MRM peptide . | Correlation between activity/antibody assay and MRM measurements . | |||
|---|---|---|---|---|---|
| Regression slope . | Pearson correlation . | R2 . | R2 95% confidence interval . | ||
| α-2 antiplasmin | LGNQEPGGQTALK | 0.68 | 0.68 | 0.46 | 0.3-0.62 |
| DFLQSLK | 0.68 | 0.68 | 0.46 | 0.31-0.63 | |
| Antithrombin-III | DDLYVSDAFHK | 0.60 | 0.67 | 0.45 | 0.25-0.66 |
| TSDQIHFFFAK | 0.71 | 0.71 | 0.50 | 0.28-0.69 | |
| FATTFYQHLADSK | 0.62 | 0.62 | 0.38 | 0.2-0.6 | |
| Coagulation factor IX | IIPHHNYNAAINK | 0.75 | 0.75 | 0.56 | 0.41-0.71 |
| SALVLQYLR | 0.74 | 0.74 | 0.55 | 0.4-0.71 | |
| VSVSQTSK | 0.46 | 0.46 | 0.21 | 0.06-0.37 | |
| Coagulation factor V | GEYEEHLGILGPIIR | 0.80 | 0.80 | 0.64 | 0.5-0.77 |
| AEVDDVIQVR | 0.89 | 0.89 | 0.80 | 0.68-0.88 | |
| SQHLDNFSNQIGK | 0.86 | 0.86 | 0.74 | 0.61-0.86 | |
| Coagulation factor VII | VAQVIIPSTYVPGTTNHDIALLR | 0.85 | 0.78 | 0.61 | 0.39-0.77 |
| Coagulation factor X | ETYDFDIAVLR | 1.08 | 0.74 | 0.55 | 0.4-0.68 |
| GYTLADNGK | 1.24 | 0.86 | 0.73 | 0.66-0.84 | |
| TGIVSGFGR | 1.26 | 0.90 | 0.81 | 0.73-0.9 | |
| Coagulation factor XI | TSESGLPSTR | 0.85 | 0.85 | 0.73 | 0.61-0.82 |
| Fibrinogen α chain | ESSSHHPGIAEFPSR | 0.68 | 0.68 | 0.46 | 0.25-0.65 |
| VQHIQLLQK | 0.74 | 0.76 | 0.58 | 0.44-0.73 | |
| GSESGIFTNTK | 0.72 | 0.72 | 0.52 | 0.29-0.71 | |
| Fibrinogen β chain | AHYGGFTVQNEANK | 0.79 | 0.79 | 0.62 | 0.43-0.77 |
| HQLYIDETVNSNIPTNLR | 0.79 | 0.79 | 0.63 | 0.46-0.76 | |
| YQISVNK | 0.71 | 0.75 | 0.57 | 0.4-0.72 | |
| Fibrinogen γ chain | IHLISTQSAIPYALR | 0.68 | 0.74 | 0.55 | 0.37-0.7 |
| YLQEIYNSNNQK | 0.76 | 0.76 | 0.58 | 0.38-0.74 | |
| YEASILTHDSSIR | 0.80 | 0.80 | 0.64 | 0.47-0.78 | |
| Plasminogen | LFLEPTR | 0.77 | 0.93 | 0.86 | 0.79-0.91 |
| Prothrombin | ELLESYIDGR | 1.34 | 0.87 | 0.76 | 0.64-0.85 |
| ETAASLLQAGYK | 1.15 | 0.78 | 0.61 | 0.46-0.76 | |
| TATSEYQTFFNPR | 1.23 | 0.85 | 0.72 | 0.54-0.85 | |
| Vitamin K–dependent protein C | DTEDQEDQVDPR | 0.88 | 0.88 | 0.77 | 0.66-0.87 |
| LGEYDLR | 0.89 | 0.90 | 0.80 | 0.72-0.87 | |
| TFVLNFIK | 0.92 | 0.92 | 0.84 | 0.75-0.91 | |
| Vitamin K–dependent protein S | IETISHEDLQR | 0.88 | 0.88 | 0.77 | 0.66-0.86 |
| SFQTGLFTAAR | 0.86 | 0.86 | 0.74 | 0.6-0.85 | |
| VYFAGFPR | 0.90 | 0.90 | 0.81 | 0.69-0.89 | |
| VWF | ILAGPAGDSNVVK | 0.11 | 0.18 | 0.03 | 0-0.26 |
| Protein . | MRM peptide . | Correlation between activity/antibody assay and MRM measurements . | |||
|---|---|---|---|---|---|
| Regression slope . | Pearson correlation . | R2 . | R2 95% confidence interval . | ||
| α-2 antiplasmin | LGNQEPGGQTALK | 0.68 | 0.68 | 0.46 | 0.3-0.62 |
| DFLQSLK | 0.68 | 0.68 | 0.46 | 0.31-0.63 | |
| Antithrombin-III | DDLYVSDAFHK | 0.60 | 0.67 | 0.45 | 0.25-0.66 |
| TSDQIHFFFAK | 0.71 | 0.71 | 0.50 | 0.28-0.69 | |
| FATTFYQHLADSK | 0.62 | 0.62 | 0.38 | 0.2-0.6 | |
| Coagulation factor IX | IIPHHNYNAAINK | 0.75 | 0.75 | 0.56 | 0.41-0.71 |
| SALVLQYLR | 0.74 | 0.74 | 0.55 | 0.4-0.71 | |
| VSVSQTSK | 0.46 | 0.46 | 0.21 | 0.06-0.37 | |
| Coagulation factor V | GEYEEHLGILGPIIR | 0.80 | 0.80 | 0.64 | 0.5-0.77 |
| AEVDDVIQVR | 0.89 | 0.89 | 0.80 | 0.68-0.88 | |
| SQHLDNFSNQIGK | 0.86 | 0.86 | 0.74 | 0.61-0.86 | |
| Coagulation factor VII | VAQVIIPSTYVPGTTNHDIALLR | 0.85 | 0.78 | 0.61 | 0.39-0.77 |
| Coagulation factor X | ETYDFDIAVLR | 1.08 | 0.74 | 0.55 | 0.4-0.68 |
| GYTLADNGK | 1.24 | 0.86 | 0.73 | 0.66-0.84 | |
| TGIVSGFGR | 1.26 | 0.90 | 0.81 | 0.73-0.9 | |
| Coagulation factor XI | TSESGLPSTR | 0.85 | 0.85 | 0.73 | 0.61-0.82 |
| Fibrinogen α chain | ESSSHHPGIAEFPSR | 0.68 | 0.68 | 0.46 | 0.25-0.65 |
| VQHIQLLQK | 0.74 | 0.76 | 0.58 | 0.44-0.73 | |
| GSESGIFTNTK | 0.72 | 0.72 | 0.52 | 0.29-0.71 | |
| Fibrinogen β chain | AHYGGFTVQNEANK | 0.79 | 0.79 | 0.62 | 0.43-0.77 |
| HQLYIDETVNSNIPTNLR | 0.79 | 0.79 | 0.63 | 0.46-0.76 | |
| YQISVNK | 0.71 | 0.75 | 0.57 | 0.4-0.72 | |
| Fibrinogen γ chain | IHLISTQSAIPYALR | 0.68 | 0.74 | 0.55 | 0.37-0.7 |
| YLQEIYNSNNQK | 0.76 | 0.76 | 0.58 | 0.38-0.74 | |
| YEASILTHDSSIR | 0.80 | 0.80 | 0.64 | 0.47-0.78 | |
| Plasminogen | LFLEPTR | 0.77 | 0.93 | 0.86 | 0.79-0.91 |
| Prothrombin | ELLESYIDGR | 1.34 | 0.87 | 0.76 | 0.64-0.85 |
| ETAASLLQAGYK | 1.15 | 0.78 | 0.61 | 0.46-0.76 | |
| TATSEYQTFFNPR | 1.23 | 0.85 | 0.72 | 0.54-0.85 | |
| Vitamin K–dependent protein C | DTEDQEDQVDPR | 0.88 | 0.88 | 0.77 | 0.66-0.87 |
| LGEYDLR | 0.89 | 0.90 | 0.80 | 0.72-0.87 | |
| TFVLNFIK | 0.92 | 0.92 | 0.84 | 0.75-0.91 | |
| Vitamin K–dependent protein S | IETISHEDLQR | 0.88 | 0.88 | 0.77 | 0.66-0.86 |
| SFQTGLFTAAR | 0.86 | 0.86 | 0.74 | 0.6-0.85 | |
| VYFAGFPR | 0.90 | 0.90 | 0.81 | 0.69-0.89 | |
| VWF | ILAGPAGDSNVVK | 0.11 | 0.18 | 0.03 | 0-0.26 |
As a fourth example, we considered antithrombin, because it is an inhibitor of a number of procoagulant serine proteases. Here, the correlations of the 3 MRM measured peptides and activity assays dropped slightly to 0.61, 0.67, and 0.71. Although the general trend was captured, these low correlations could possibly be because antithrombin was measured by a multistep activity assay.
The next example is the anticoagulant protein C, for which we observed good correlations for all 3 peptides (0.87, 0.89, and 0.91) and, as expected, low extreme values for samples from patients using vitamin K antagonist medication.
Plasminogen is also included in the examples represented in Figure 2. We observed a high correlation of 0.93 of the 1 peptide included (ie, LFLEPTR) with the activity assay for that protein.
The last example is protein S, which circulates in the blood as a complex with C4b binding protein. Therefore, although C4b binding protein was not determined by activity or antibody assays in this study, its level measured by MRM correlated with the protein S antibody assay well, as expected, with a Pearson correlation of 0.82.
Table 1 summarizes the correlations of all 36 measured peptides with their corresponding antibody or activity assay protein levels, represented by Pearson correlation coefficients as well as explained variance R2 values. Most regression slopes are <1, with an average of 0.81, which means that the MRM measurements (x-axis) were slightly more sensitive than antibody/activity assay measurements (y-axis; Figure 2; Table 1). A histogram of the Pearson correlation coefficient values showed that a majority of protein concentration levels determined in both approaches correlated well, with an average of 0.77, with 23 of the 36 correlations >0.75. The 1 poorly performing MRM measurement, with many missing values, concerned VWF (Table 1).
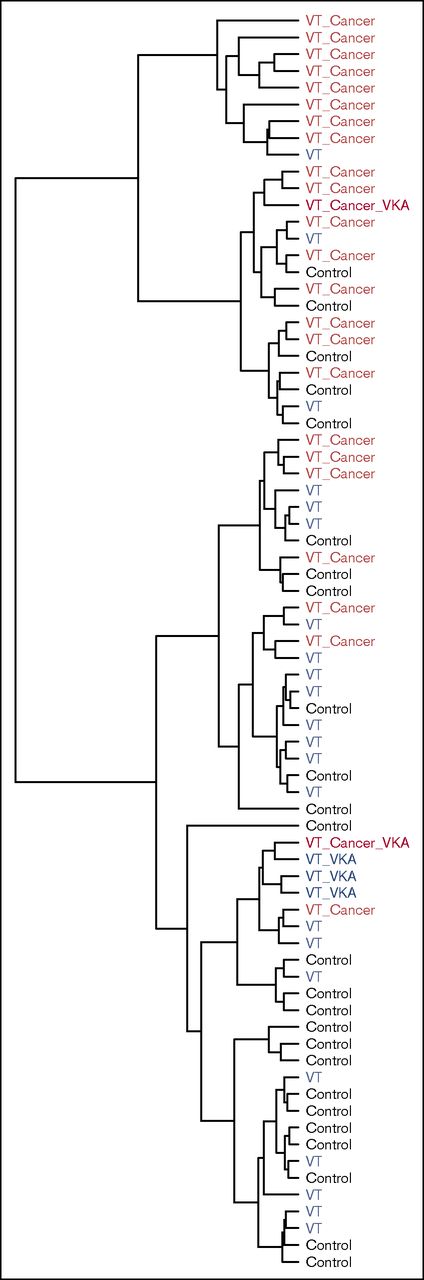
Unsupervised cluster analysis
Although the main goal of the study was to investigate the correlation between MRM and activity/antibody assay, we applied an unsupervised hierarchal cluster analysis of the MRM data to examine any underlying grouping in the measured samples. After sharing the metadata on the individual samples, we found out that the cluster analysis grouped 17 of 25 patients with VT and cancer in 1 cluster, meaning that there was a shared signature in the collected samples of these patients (Figure 3). Not surprisingly, 4 of the 5 patients receiving vitamin K antagonist medication at the time of blood collection clustered together.
Unsupervised hierarchical cluster analysis of the peptide concentration levels measured by LC-MRM/MS of the 75 samples. The analysis was able to discriminate 17 of 25 patients with thrombosis and cancer. VKA, vitamin K antagonist.
Unsupervised hierarchical cluster analysis of the peptide concentration levels measured by LC-MRM/MS of the 75 samples. The analysis was able to discriminate 17 of 25 patients with thrombosis and cancer. VKA, vitamin K antagonist.
To compare more closely the MRM versus activity/antibody assays, we performed the clustering again using only the 13 proteins measured in both approaches (supplemental Table 2). MRM proved to be the better discriminator by grouping 16 of the 25 patients with VT and cancer, whereas the measurements from activity/antibody assays were scattered, with a maximum of 10 in 1 cluster. This shows that coagulation factor levels measured by MRM carry signature information on thrombosis and cancer, which is not obvious from a single measurement.
Discussion
We performed the first MS measurement in a case-control study of 75 patients. We measured 31 proteins, of which 13 had been assayed by antibody or activity assay previously, in a sample from the same blood draw. For all but 1 protein (VWF), we found high correlations between the MS and traditional assays, with correlation coefficients ranging from 0.62 to 0.93 and averaging 0.77. By cluster analysis, we were able to discriminate clearly patients with cancer and thrombosis from those with only thrombosis or neither.
Profiling of blood plasma protein levels is a cornerstone of diagnosis and etiologic research. Currently, antibody and activity assays are most commonly used for this purpose. However, because of their low multiplicity, relatively high cost, and required sample volume, it is challenging to perform studies with large sample sizes. Recently, VT-associated proteins were also evaluated using affinity proteomics based on suspension-bead arrays for discovery followed by immunocapture MS for validation,24 which, similar to antibody assays, depends mainly on the availability of ≥1 antibody per target protein.
In our study, we compared the conventional antibody and activity assay measurements to determine plasma protein content with the MS approach of targeted proteomic MRM using internal heavy-labeled standards. Although the antibody and activity assay measurements were collected over time using 1 assay per protein, our MS approach exploited the multiplicity power of targeted proteomics to measure 31 proteins in 1 run for each sample. All LC-MRM/MS measurements of all samples and targets were accomplished in <4 days, including the preparation of the internal standard mixture used, and only 3 μL per sample was required to analyze the 31 target proteins.
When comparing the conventional and the MS MRM strategies, a mean Pearson correlation for the 13 proteins (covered by 36 target peptides) shared between the 2 approaches was 0.77, indicating a good and promising correlation between the 2 strategies. In addition, higher sensitivity in MRM (mean regression slope, 0.81), higher multiplicity in a single run (19 additional proteins from the coagulation and related pathways), and good ability to leverage all measurements to discriminate between groups (unsupervised clustering of groups of vitamin K antagonist users as well as patients with VT and cancer) imply that LC-MRM/MS could be used as a good alternative to conventional methods.
In terms of quality of MRM, we observed absence of or a poor signal for a few proteins, notably VWF. It is unclear whether this was because of digestion efficiency, sample handling, protein degradation resulting from intrinsic plasma proteases and peptidases (especially because the samples were anticoagulated using citrate, not a stronger protease inhibiter like K2-EDTA), or possibly instrument/scheduling issues. The MRM measurements were performed in a single-step procedure with no balancing of the SIS peptides to the endogenous peptides22 or checking of the detectability of endogenous peptides or whether there was interference in these specific samples (or pool thereof); therefore, improvement is possible. It important to note that MRM, like most MS methods, relies on standardized sample handling to ensure valid comparison between samples. Although we have not demonstrated the reproducibility of our MRM assay on the 75 samples included in this work, several previous studies in other fields have reported high reproducibility.25,26 Compared with activity assays, MRM seems to have an inherited shortcoming because it does not allow the assessment of protein activity. However, the 2 technologies showed good correlation for most proteins and can be considered complementary, for example, for evaluating the activity as well as the abundance of vitamin K–dependent factors in a cohort with patients receiving vitamin K antagonist medication. Such cases were visible as outliers (Figure 1), and MRM and activity assays still showed good correlation. An advantage of MRM is that it is a targeted method, which allows a specific measurement, as, for example, in the case of the activation peptide of vitamin K–dependent protein C (supplemental Report 1; peptide DTEDQEDQVDPR).
With unsupervised clustering analysis, 17 of the 25 individuals experiencing VT with associated cancer were grouped together in the largest cluster. The same clustering using the activity and antibody assay data grouped only 10 in distinct clusters associated with these conditions. These data show that a combination of coagulation factor levels yields signature information on thrombosis and cancer, which is not obvious from single measurements and may offer diagnostic potential.
The emphasis in this study was to test the feasibility of multiplexed profiling of protein levels of coagulation factors to be applied in large cohort. The results encourage the further validation and investigation of MRM as an alternative to conventional assays in profiling protein signature of disease.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Ingeborg de Jonge and Petra Noordijk for their help and support in sample collection and preparation and Suzanne C. Cannegieter, Henri H. Versteeg, and Pieter H. Reitsma for the fruitful discussions.
This work was supported by Genome Canada and Genome British Columbia (project codes 204PRO for operations and 214PRO for technology development). C.H.B. is grateful for support from the Leading Edge Endowment Fund (University of Victoria), the Segal McGill Chair in Molecular Oncology at McGill University (Montreal, QC, Canada), the Warren Y. Soper Charitable Trust, and the Alvin Segal Family Foundation contribution to the Jewish General Hospital (Montreal, QC, Canada).
Authorship
Contribution: F.R.R. initiated the project, collected the samples, and performed the activity/antibody analyses; Y.M., B.J.v.V., M.P., C.H.B., and F.R.R. planned the MRM experiment; Y.M., J.Y., A.J.P., and C.H.B. conduced the MRM analyses; Y.M. performed the data analysis, correlation, and unsupervised clustering; Y.M., B.J.v.V., and F.R.R. reviewed the results and wrote the initial draft of the manuscript; and all authors approved the manuscript.
Conflict-of-interest disclosure: C.H.B. is the chief strategy officer of MRM Proteomics, Inc. The remaining authors declare no competing financial interests.
Correspondence: Frits R. Rosendaal, Department of Clinical Epidemiology, Leiden University Medical Center, P.O. Box 9600, 2300 RC Leiden, The Netherlands; e-mail: f.r.rosendaal@lumc.nl.
References
Author notes
Y.M. and B.J.v.V. contributed equally to this work.