Key Points
MLOF used an innovative approach to genotype 3000 hemophilia patients identifying likely causative variants in 98.4% of patients.
Hemophilia genotyping should include structural variation, F8 inversions (for hemophilia A), and consideration of gene-wide approaches.
Abstract
Hemophilia A and B are rare, X-linked bleeding disorders. My Life, Our Future (MLOF) is a collaborative project established to genotype and study hemophilia. Patients were enrolled at US hemophilia treatment centers (HTCs). Genotyping was performed centrally using next-generation sequencing (NGS) with an approach that detected common F8 gene inversions simultaneously with F8 and F9 gene sequencing followed by confirmation using standard genotyping methods. Sixty-nine HTCs enrolled the first 3000 patients in under 3 years. Clinically reportable DNA variants were detected in 98.1% (2357/2401) of hemophilia A and 99.3% (595/599) of hemophilia B patients. Of the 924 unique variants found, 285 were novel. Predicted gene-disrupting variants were common in severe disease; missense variants predominated in mild–moderate disease. Novel DNA variants accounted for ∼30% of variants found and were detected continuously throughout the project, indicating that additional variation likely remains undiscovered. The NGS approach detected >1 reportable variants in 36 patients (10 females), a finding with potential clinical implications. NGS also detected incidental variants unlikely to cause disease, including 11 variants previously reported in hemophilia. Although these genes are thought to be conserved, our findings support caution in interpretation of new variants. In summary, MLOF has contributed significantly toward variant annotation in the F8 and F9 genes. In the near future, investigators will be able to access MLOF data and repository samples for research to advance our understanding of hemophilia.
Introduction
Hemophilia A and B are X-linked recessive disorders resulting from more than 3000 different DNA variants reported to date in the genes encoding coagulation factor VIII (FVIII) and FIX, respectively.1-5 Determination of the causative genetic variant in families affected by hemophilia is important for use in reproductive planning, for use in pregnancy and neonatal management, and also to inform risks of neutralizing antibody (inhibitor) formation and bleeding severity.6-12 Therapies targeted to specific mutations have been studied and are likely to become more common in the future.13
In 2012, 2 separate surveys, 1 distributed to hemophilia providers through the American Thrombosis Hemostasis Network (ATHN) and the other distributed to the patient community through the National Hemophilia Foundation, found that only ∼20% of the hemophilia patients in the United States had had their genotype determined. The most common reasons cited for not having the testing performed were the lack of medical insurance coverage and the lack of availability of testing. Furthermore, the scientific and clinical communities recognized unmet research needs in hemophilia diagnosis, mechanisms, complications, and treatment.
The My Life, Our Future (MLOF) project is a formal, multisector collaboration among ATHN, National Hemophilia Foundation, Bloodworks Northwest (BWNW) (formerly the Puget Sound Blood Center), and Bioverativ. It was developed to provide wide-scale access to free hemophilia genotype analysis for patients in the United States and to create a research repository of associated samples and data to support scientific discovery and treatment advances.
In this manuscript, we describe our novel genotyping approach and report the results from the first 3000 hemophilia A and B patients enrolled in the project, representing ∼15% of the total hemophilia A and B population in the United States.
Materials and methods
Patient enrollment
Participating hemophilia treatment center (HTC) providers contracted through ATHN to enroll patients, obtained samples for genotyping, and provided clinical results to their patients. Patients were required to have a diagnosis of hemophilia A or B and a documented FVIII or FIX level <50% at baseline. The project began with a pilot involving 11 HTCs and subsequently was offered to the rest of the US HTC network. BWNW served as the central genotyping laboratory.
F8 and F9 gene variant analysis
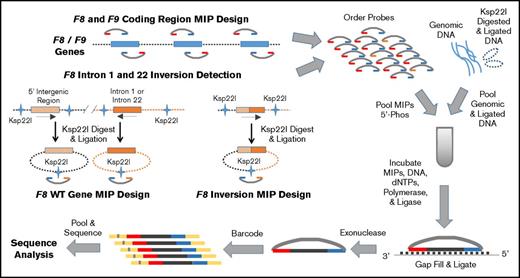
DNA was extracted from EDTA anticoagulated blood using whole blood DNA extraction kits and automated technology (Gentra Puregene Blood Kit and QIAsymphony; Qiagen Inc., Germantown, MD). Initial variant analysis was performed at the University of Washington utilizing an F8 and F9 gene-targeted next-generation sequencing (NGS) approach, which employed molecular inversion probes (MIPs) for DNA capture14-17 (see Figure 1). Briefly, the MLOF MIPs are single-stranded custom DNA molecules that were designed to have a common internal linker sequence flanked 5′ and 3′ by sequences complementary to genomic target regions, which in MLOF were usually 111 bp in size. MIPs were annealed to genomic DNA, the gap filled by a polymerase using the genomic DNA as a template, ends ligated to circularize the probe, and probes containing the complementary genomic target sequences released by exonuclease digestion. Barcoding of linearized probes permitted pooling of samples for NGS and enabled high sample throughput. Taken together, the MIP targets capture all F8 and F9 coding regions, splice sites, and upstream (450 bp for F8 and 300 bp for F9) and downstream (1838 bp for F8 and 1417 bp for F9) untranslated sequences determined by the complementary DNA (cDNA) sequences NM_000132.3 for F8 and NM_000133.3 for F9 (see supplemental Table 1 for MIP sequences). Use of this targeted sequencing strategy resulted in sequencing of both the F8 and F9 genes in all patients.
MLOF F8 and F9 MIP next-generation DNA sequencing strategy. Schematic of the MIP targeted sequencing of the F8 and F9 genes in hemophilia. First, MIPs were designed to capture all coding, 5′, and 3′ untranslated regions (UTRs) of both the F8 and F9 genes. Additional MIPs were designed to detect unique sequences produced when genomic DNA carrying one of the common F8 intron 1 or intron 22 inversions is digested with Ksp22I and ligated as well as the normal reference, or wild-type, F8 sequence. Genomic and Ksp22I digested and ligated DNA was prepped and mixed with pooled dephosphorylated MIPs, 2′-deoxynucleoside 5'-triphosphates (dNTPs), polymerase, and ligase for MIP target gap filling and ligation. MIPs were released by exonuclease digestion, and the library was individually bar-coded, pooled, and sequenced using an Illumina MiSeq or NextSeq (schematic adapted from O’Roak et al16 ). The resulting DNA sequence data were cleaned, subjected to quality control filters, aligned to the reference genome (hg37), and annotated for analysis in the clinical laboratory.
MLOF F8 and F9 MIP next-generation DNA sequencing strategy. Schematic of the MIP targeted sequencing of the F8 and F9 genes in hemophilia. First, MIPs were designed to capture all coding, 5′, and 3′ untranslated regions (UTRs) of both the F8 and F9 genes. Additional MIPs were designed to detect unique sequences produced when genomic DNA carrying one of the common F8 intron 1 or intron 22 inversions is digested with Ksp22I and ligated as well as the normal reference, or wild-type, F8 sequence. Genomic and Ksp22I digested and ligated DNA was prepped and mixed with pooled dephosphorylated MIPs, 2′-deoxynucleoside 5'-triphosphates (dNTPs), polymerase, and ligase for MIP target gap filling and ligation. MIPs were released by exonuclease digestion, and the library was individually bar-coded, pooled, and sequenced using an Illumina MiSeq or NextSeq (schematic adapted from O’Roak et al16 ). The resulting DNA sequence data were cleaned, subjected to quality control filters, aligned to the reference genome (hg37), and annotated for analysis in the clinical laboratory.
Approximately 50% of severe hemophilia A (FVIII <1%) has been attributed to large DNA inversions mediated through sequences in F8 intron 22 (∼45%) or F8 intron 1 (2% to 5%) and homologous sequences distal to the F8 gene, which result in disruption of the F8 gene.7,18,19 To detect these common F8 inversions, MIPs were designed to capture ligated mutant or reference sequences using an approach similar to the inverse-shifting polymerase chain reaction (PCR) methodology described by Rossetti et al.20 Briefly, 500 ng of genomic DNA was cleaved by Ksp22I (SibEnzyme US LLC, West Roxbury, MA) (1 hour at 37°C), followed by heat inactivation (20 minutes at 65°C), and subsequently ligated with T4 Ligase (New England Biolabs Inc., Ipswich, MA) (16 hours at 15°C) followed by heat inactivation (10 minutes at 65°C). The Ksp221 digested/ligated product (25 ng) was combined with 100 ng of unmodified genomic DNA prior to PCR amplification and MIP capture (Figure 1). High-throughput DNA sequencing by synthesis was performed using MiSeq or NextSeq instruments (Illumina Inc., San Diego, CA).
For sequence analysis, following removal of MIP arms, a joint InDel realignment was performed (Broad Institute’s Genome Analysis Tool Kit 3.2-2), and subsequently, variants for each run position were called using UnifiedGenotyper (Broad Institute’s Genome Analysis Tool Kit 3.4-46). Sequences were aligned to the human reference genome (GRCh37 1000 Genomes Phase II release) and inversion reference sequences using bwa-0.7.5 MEM (http://bio-bwa.sourceforge.net/). Ensembl Variant Effect Predictor (Ensembl release 76; http://www.ensembl.org/info/docs/tools/vep/script/index.html) was used with the last GRCh37 annotation build (Ensembl release 75) to annotate variants.
Variants identified by NGS were confirmed in the BWNW Clinical Laboratory Improvement Amendments–certified clinical genomics laboratory by a method specific to the variant using a second sample aliquot. Methods included gel analysis of restriction enzyme fragment cleavage products of a genomic PCR amplified product, genomic PCR amplification, Sanger sequencing (ABI BigDye Terminator v3.1; ABI 3130xl Genetic Analyzer; Life Technologies, Grand Island, NY) or, for F8 inversions, inverse-shifting PCR (intron 1) or long-range PCR (intron 22).20,21 In individuals with no likely deleterious variant identified by NGS, Sanger sequencing of the coding, splice sites, and immediate upstream and downstream regions, similar but not identical in genomic coverage to the NGS MIP targets, was performed. In patients reported to have moderate or severe hemophilia A without a variant detected, inverse-shifting and long-range PCR was performed to further exclude intron 22 and intron 1 inversions. In males in whom no likely deleterious variant was identified, females with moderate or severe disease with either no variant or only 1 likely deleterious variant identified, and subjects in whom NGS read depth suggested large structural variation that was not validated by another method, multiplex ligation-dependent probe amplification (F8-178 and F9-207 kits; MRC-Holland, Amsterdam, the Netherlands) was used to assess for structural variants (SVs) in the candidate gene.
F8 and F9 sequences were aligned using Mutation Surveyor v.4.0 (SoftGenetics LLC, State College, PA), and identified variants were compared with those published in the European Association for Haemophilia and Allied Disorders (EAHAD) Coagulation Factor Variant databases (http://www.factorviii-db.org; http://www.factorix.org),1,2 the Centers for Disease Control and Prevention (CDC) Hemophilia Mutation Project databases (http://www.cdc.gov/ncbddd/hemophilia/champs.html),3,4 the Spanish Hemobase (http://www.hemobase.com/EN/index.htm),5 and those found previously in the BWNW laboratory.
The BWNW clinical genomics laboratory is a laboratory with historical expertise in interpreting genetic testing data in hemophilia. The BWNW laboratory determined clinical actionability of F8 and F9 DNA variants for validation and clinical reporting per the laboratory’s hemophilia genotype annotation. DNA variants previously reported by others or known to our laboratory to be benign variants (polymorphisms) and variants within the region encoding the FVIII B domain not previously shown to impact F8 gene function were excluded. Clinical interpretation accounts for gender, disease (hemophilia A or B), assigned disease severity using reported baseline factor level, variant location, prior reporting of the variant in hemophilia, and, when available, genetic evidence of segregation of the variant with hemophilia in families, genetic evidence that a variant arose de novo, and published in vitro functional data (see supplemental Table 2 for a list of F8 and F9 DNA variants not reported). Beginning in July 2014, clinical interpretation used criteria for pathogenicity published by the American College of Medical Genetics and Genomics.22
Results
Subject enrollment and characteristics
Sixty-nine HTCs enrolled the first 3000 patients, beginning with 11 pilot sites (see supplemental Table 3). In the first 3000 participants, 2401 patients had hemophilia A and 599 patients had hemophilia B. The distribution of MLOF participants by hemophilia type, severity, and sex is shown in Table 1. Patients with a diagnosis of hemophilia A or B of all severities, both males and females, were eligible for the project. The diagnosis of hemophilia was established by the local HTC.
MLOF patient demographics
| Hemophilia type . | Number of males . | Number by severity (severe/moderate/mild) . | Number of females . | Number by severity (severe/moderate/mild) . | Total . |
|---|---|---|---|---|---|
| A | 2320 | 1272/441/6073 | 81 | 8/7/66 | 2401 |
| B | 580 | 217/222/141 | 19 | 0/0/19 | 599 |
| Total | 2900 | 1489/663/748 | 100 | 8/7/85 | 3000 |
| Hemophilia type . | Number of males . | Number by severity (severe/moderate/mild) . | Number of females . | Number by severity (severe/moderate/mild) . | Total . |
|---|---|---|---|---|---|
| A | 2320 | 1272/441/6073 | 81 | 8/7/66 | 2401 |
| B | 580 | 217/222/141 | 19 | 0/0/19 | 599 |
| Total | 2900 | 1489/663/748 | 100 | 8/7/85 | 3000 |
F8 and F9 variant analysis
In an analysis of the 3000 patients, clinically reportable genetic variation was detected in 98.1% (2357/2401) of patients with hemophilia A and 99.3% (595/599) of patients with hemophilia B. In 48 individuals (44 males), no potentially causative DNA variant was found by NGS, Sanger sequencing, F8 inversion detection by long-range and inverse-shifting PCR, and multiplex ligation-dependent probe amplification. Of these, 44 had been diagnosed with hemophilia A (6 females: 1 severe and 5 mild; 38 males: 8 severe, 4 moderate, and 26 mild), and 4 had been diagnosed with hemophilia B (4 males: 1 moderate and 3 mild) (distributions of disease severity in MLOF are in Table 1).
Overall, 100 women were enrolled, 81 with hemophilia A (8 severe, 7 moderate, and 66 mild) and 19 with hemophilia B (all mild). Clinically reportable DNA variants were detected in 75/81 (93%) of women with hemophilia A and 19/19 (100%) of women with hemophilia B.
More than 1 potentially causative DNA variant in F8 was detected in 35 patients with hemophilia A. We did not find more than 1 potentially causative variant in F9 in patients with hemophilia B. Of patients with more than 1 variant, 25 were males (22 severe, 1 moderate, and 2 mild; see Table 2) whose variants were presumed to lie in cis, and 10 were females (2 severe, 1 moderate, and 7 mild; see Table 3) for whom variants could lie in cis or trans.
Male subjects in whom 2 reportable variants were detected (all with hemophilia A)
| Baseline level (%) . | First variant HGVS cDNA . | First variant exon/intron . | First variant HGVS protein . | Second variant HGVS cDNA . | Second variant exon/intron . | Second variant HGVS protein . |
|---|---|---|---|---|---|---|
| <1* | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6188-?_6429+?del | Exons 21-22 | (Deletion) |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.1908delGinsCATCAAAGTACTTCAAAAA | Exon 17 | p.Trp1908delinsSerSerLysTyrPheLysLys |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.-279C>T | 5′UTR | NA |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6929C>T | Exon 26 | p.Thr2291Ile |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.1-?_6429+?del | Exons 1-22 | (Deletion) |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6430-?_6900+?dup | Exons 23-25 | (Duplication) |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6116-?_6429+?del | Exons 20-22 | (Deletion) |
| <1 | c.1139A>G | Exon 8 | p.Asp380Gly | c.5999-3_6002del | Intron 18/exon 19 | Gly2000Valfs*29† |
| <1 | c.1313T>C‡ | Exon 9 | p.Ile438Thr | c.1373G>A‡ | Exon 9 | Arg458His |
| <1 | c.144-?_601+?dup§ | Exons 2-4 | (Duplication) | c.671-?_787+?dup§ | Exon 6 | (Duplication) |
| <1 | c.1538-18G>A§ | Intron 10 | (Splice) | c.1538-13delT§ | Intron 10 | (Splice) |
| <1 | c.311T>G | Exon 3 | p.Val104Gly | c.343G>C | Intron 3 | p.Val115Leu |
| <1 | c.389-?_1443+?del | Exons 4-9 | (Deletion) | c.1538-?_1903+?del | Exons 11-12 | (Deletion) |
| <1 | c.5705T>G | Exon 17 | p.Phe1902Cys | c.5725T>C | Exon 17 | p.Tyr1909His |
| <1 | c.575T>C | Exon 4 | p.Ile192Thr | c.589G>C | Exon 4 | p.Val197Leu |
| <1 | c.5901C>G | Exon 18 | p.(=) | c.5921C>A | Exon 18 | p.Ser1974Tyr |
| <1 | c.6406C>T | Exon 19 | p.Arg2016Trp | c.6724G>A | Exon 25 | p.Val2242Met |
| <1 | c.6046C>T | Exon 19 | p.Arg2016Trp | c.6403C>T | Exon 22 | p.Arg2135* |
| 2 | c.5247C>G | Exon 15 | p.Phe1749Leu | c.5302C>T | Exon 15 | Arg1768Cys |
| 8 | c.1280A>T | Exon 9 | p.Lys427Ile | c.1309C>T | Exon 9 | p.Arg437Trp |
| 22 | c.655G>A | Exon 5 | p.Ala219Thr | c.6583C>A | Exon 24 | p.Met2195Leu |
| Baseline level (%) . | First variant HGVS cDNA . | First variant exon/intron . | First variant HGVS protein . | Second variant HGVS cDNA . | Second variant exon/intron . | Second variant HGVS protein . |
|---|---|---|---|---|---|---|
| <1* | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6188-?_6429+?del | Exons 21-22 | (Deletion) |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.1908delGinsCATCAAAGTACTTCAAAAA | Exon 17 | p.Trp1908delinsSerSerLysTyrPheLysLys |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.-279C>T | 5′UTR | NA |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6929C>T | Exon 26 | p.Thr2291Ile |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.1-?_6429+?del | Exons 1-22 | (Deletion) |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6430-?_6900+?dup | Exons 23-25 | (Duplication) |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | c.6116-?_6429+?del | Exons 20-22 | (Deletion) |
| <1 | c.1139A>G | Exon 8 | p.Asp380Gly | c.5999-3_6002del | Intron 18/exon 19 | Gly2000Valfs*29† |
| <1 | c.1313T>C‡ | Exon 9 | p.Ile438Thr | c.1373G>A‡ | Exon 9 | Arg458His |
| <1 | c.144-?_601+?dup§ | Exons 2-4 | (Duplication) | c.671-?_787+?dup§ | Exon 6 | (Duplication) |
| <1 | c.1538-18G>A§ | Intron 10 | (Splice) | c.1538-13delT§ | Intron 10 | (Splice) |
| <1 | c.311T>G | Exon 3 | p.Val104Gly | c.343G>C | Intron 3 | p.Val115Leu |
| <1 | c.389-?_1443+?del | Exons 4-9 | (Deletion) | c.1538-?_1903+?del | Exons 11-12 | (Deletion) |
| <1 | c.5705T>G | Exon 17 | p.Phe1902Cys | c.5725T>C | Exon 17 | p.Tyr1909His |
| <1 | c.575T>C | Exon 4 | p.Ile192Thr | c.589G>C | Exon 4 | p.Val197Leu |
| <1 | c.5901C>G | Exon 18 | p.(=) | c.5921C>A | Exon 18 | p.Ser1974Tyr |
| <1 | c.6406C>T | Exon 19 | p.Arg2016Trp | c.6724G>A | Exon 25 | p.Val2242Met |
| <1 | c.6046C>T | Exon 19 | p.Arg2016Trp | c.6403C>T | Exon 22 | p.Arg2135* |
| 2 | c.5247C>G | Exon 15 | p.Phe1749Leu | c.5302C>T | Exon 15 | Arg1768Cys |
| 8 | c.1280A>T | Exon 9 | p.Lys427Ile | c.1309C>T | Exon 9 | p.Arg437Trp |
| 22 | c.655G>A | Exon 5 | p.Ala219Thr | c.6583C>A | Exon 24 | p.Met2195Leu |
HGVS, Human Genome Variation Society; NA, not available.
Subject also had c.2114-?_5219+?_del, which results in deletion of exon 14.
If exon 19 is transcribed.
Found in 3 subjects with same 3 variants.
Found in 2 subjects with same 2 variants.
Female subjects in whom 2 reportable variants were detected (all with hemophilia A)
| Female baseline level (%) . | First variant HGVS cDNA . | First variant exon/intron . | First variant HGVS protein . | Reported male severity* . | Second variant HGVS cDNA . | Second variant exon/intron . | Second variant HGVS protein . | Reported male severity* . |
|---|---|---|---|---|---|---|---|---|
| 12 | c.1-?_143+del | Exon 1 | (Deletion) | Moderate, severe | c.6274-?_6429+?del | Exon 22 | (Deletion) | Moderate, severe1 |
| <1 | c.143+?_144-?inv | Intron 1 | (F8 inversion) | Severe | c.144-?_601+?del | Exons 2-4 | (Deletion) | Severe1 |
| 26 | c.1302C>T | Exon 9 | p.(=) | NA | c.1331_1333delinsT | Exon 9 | p.Lys444ilefs*9 | NA |
| 29 | c.1834C>T | Exon 12 | p.Arg612Cys | Mild, moderate | c.6622C>G | Exon 24 | p.Gln2208Glu | Mild |
| 37 | c.2167G>A | Exon 14 | p.Ala723Thr | Mild, moderate, severe | c.6066C>G | Exon 19 | p.(=) | NA |
| 15 | c.2167G>A | Exon 14 | p.Ala723Thr | Mild, moderate, severe | c.6871A>G | Exon 25 | p.Thr2291Ala | NA |
| 1 | c.5878C>A | Exon 18 | p.(=) | Moderate, severe | c.1538-2A>G | Intron 10 | (Splice) | Severe1 |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | Severe | c.1-?_6429+?del | Exons 1-22 | (Deletion) | Severe1†,‡ |
| 18 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | Severe | c.6430-?_6900+?dup | Exons 23-25 | (Duplication) | Severe† |
| 34 | c.6632C>T | Exon 24 | p.Ala2211Val | Mild | c.6929C>T | Exon 26 | p.Thr2310Ile | Mild |
| Female baseline level (%) . | First variant HGVS cDNA . | First variant exon/intron . | First variant HGVS protein . | Reported male severity* . | Second variant HGVS cDNA . | Second variant exon/intron . | Second variant HGVS protein . | Reported male severity* . |
|---|---|---|---|---|---|---|---|---|
| 12 | c.1-?_143+del | Exon 1 | (Deletion) | Moderate, severe | c.6274-?_6429+?del | Exon 22 | (Deletion) | Moderate, severe1 |
| <1 | c.143+?_144-?inv | Intron 1 | (F8 inversion) | Severe | c.144-?_601+?del | Exons 2-4 | (Deletion) | Severe1 |
| 26 | c.1302C>T | Exon 9 | p.(=) | NA | c.1331_1333delinsT | Exon 9 | p.Lys444ilefs*9 | NA |
| 29 | c.1834C>T | Exon 12 | p.Arg612Cys | Mild, moderate | c.6622C>G | Exon 24 | p.Gln2208Glu | Mild |
| 37 | c.2167G>A | Exon 14 | p.Ala723Thr | Mild, moderate, severe | c.6066C>G | Exon 19 | p.(=) | NA |
| 15 | c.2167G>A | Exon 14 | p.Ala723Thr | Mild, moderate, severe | c.6871A>G | Exon 25 | p.Thr2291Ala | NA |
| 1 | c.5878C>A | Exon 18 | p.(=) | Moderate, severe | c.1538-2A>G | Intron 10 | (Splice) | Severe1 |
| <1 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | Severe | c.1-?_6429+?del | Exons 1-22 | (Deletion) | Severe1†,‡ |
| 18 | c.6429+?_6430-?inv | Intron 22 | (F8 inversion) | Severe | c.6430-?_6900+?dup | Exons 23-25 | (Duplication) | Severe† |
| 34 | c.6632C>T | Exon 24 | p.Ala2211Val | Mild | c.6929C>T | Exon 26 | p.Thr2310Ile | Mild |
NA, not applicable because of lack of hemophilia severity information in males.
Unless otherwise indicated, the data on male severity associated with each variant are from MLOF.
Variant found in a male with severe hemophilia who also had the other female reported variant.
In EAHAD database (ref#1), c.1-?_6429+?del is reported associated with severe hemophilia in females.
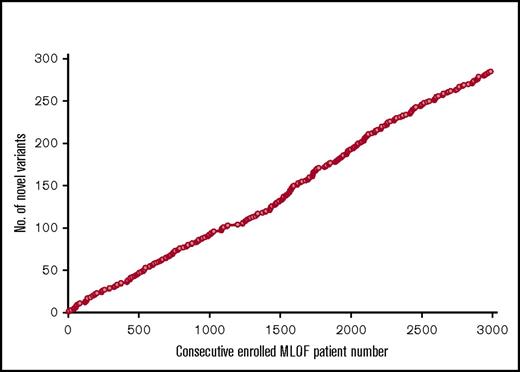
A total of 924 unique potentially causative DNA variants were found, 707 in F8 and 217 in F9. Of these, 285 variants were novel (230 in F8 and 55 in F9), defined as not having been reported in and absent from the CDC,3,4 EAHAD,1,2 and Hemobase5 databases prior to detection in MLOF. Unique novel variants continued to be detected throughout the project (Figure 2). These variants will be reported in the CDC and EAHAD databases and are also reported here in supplemental Table 4. Novel variants were sometimes detected more than once in MLOF, with a total of 334 patients (269 with hemophilia A and 65 with hemophilia B) found to have previously unknown F8 or F9 gene variants.
Detection of 385 unique novel variants in 3000 MLOF patients with hemophilia throughout the project. The totals of first-time-detected unique novel F8 and F9 variants in MLOF are shown vs the enrollment number of the MLOF subjects.
Detection of 385 unique novel variants in 3000 MLOF patients with hemophilia throughout the project. The totals of first-time-detected unique novel F8 and F9 variants in MLOF are shown vs the enrollment number of the MLOF subjects.
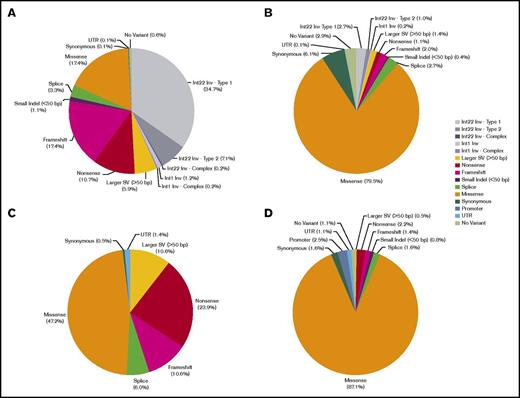
The frequency of different DNA variant types (eg, missense, nonsense, etc.) are shown in Figure 3 (and supplemental Table 5) by male hemophilia disease severity. Missense variants accounted for most of the variants detected in males with mild or moderate hemophilia A or B (79.5% in F8, 87.1% in F9). Nonsense, frameshift, and larger (>50 bp) SVs including inversions made up the majority of variants detected in males with severe hemophilia A (77.4%) and a large proportion of the variants detected in males with severe hemophilia B (45.0%), consistent with the predicted negative impact of these types of variants on gene function. The locations of variants relative to the F8 and F9 gene coding regions is shown in Figure 4 for SNVs by male hemophilia severity and for large (>50 bp) non-F8 inversion SVs (deletions, duplications, and insertions).
Frequencies of different types of F8 and F9 DNA variants detected in hemophilia. Classification of DNA variants detected in males with severe hemophilia A (A), mild–moderate hemophilia A (B), severe hemophilia B (C), and mild–moderate hemophilia B (D). For both hemophilia A and B, structural and nonsense variants predominate in severe disease, whereas missense variants account for most variants detected in mild–moderate disease.
Frequencies of different types of F8 and F9 DNA variants detected in hemophilia. Classification of DNA variants detected in males with severe hemophilia A (A), mild–moderate hemophilia A (B), severe hemophilia B (C), and mild–moderate hemophilia B (D). For both hemophilia A and B, structural and nonsense variants predominate in severe disease, whereas missense variants account for most variants detected in mild–moderate disease.
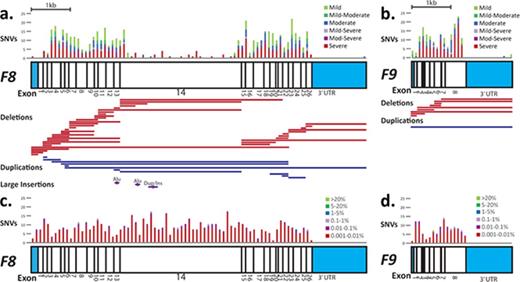
Schematic of the locations of single nucleotide substitutions (single nucleotide variants [SNVs]), excluding nonsense variants; SVs in hemophilia A and B; and Exome Aggregation Consortium (ExAC) database SNVs relative to the F8 and F9 cDNAs. Scale schematics of the coding regions of the F8 gene (A, C) and the F9 gene (B, D). UTRs are depicted with blue filled boxes; exons are depicted as open boxes and numbered beneath. In panels A and B, above each gene, the frequency of unique SNV substitutions (missense, synonymous, splice, and UTR) by gene location is shown for males, and stacked histograms are colored by hemophilia disease severity (red, severe; purple, moderate–severe; light purple, mild–severe; blue, moderate; teal, mild–moderate; green, mild); histograms bins are 100 nucleotides. Below each gene, regions impacted by unique SVs are shown (red bars indicate deletions, blue bars indicate duplications, and pink hexagons indicate insertions). In panels C and D, above each gene, the incidence of unique SNV substitutions reported in the ExAC database of unrelated individuals is shown by gene location, and stacked histograms are colored by ExAC population frequency (red, 0.001% to 0.01%; purple, 0.01% to 0.1%; light purple, 0.1% to 1%; blue, 1% to 5%; teal, 5% to 20%; green, >20%); histogram bins are 100 nucleotides. For both genes, there was no annotated variation reported in the 3′UTRs after position c.*52.
Schematic of the locations of single nucleotide substitutions (single nucleotide variants [SNVs]), excluding nonsense variants; SVs in hemophilia A and B; and Exome Aggregation Consortium (ExAC) database SNVs relative to the F8 and F9 cDNAs. Scale schematics of the coding regions of the F8 gene (A, C) and the F9 gene (B, D). UTRs are depicted with blue filled boxes; exons are depicted as open boxes and numbered beneath. In panels A and B, above each gene, the frequency of unique SNV substitutions (missense, synonymous, splice, and UTR) by gene location is shown for males, and stacked histograms are colored by hemophilia disease severity (red, severe; purple, moderate–severe; light purple, mild–severe; blue, moderate; teal, mild–moderate; green, mild); histograms bins are 100 nucleotides. Below each gene, regions impacted by unique SVs are shown (red bars indicate deletions, blue bars indicate duplications, and pink hexagons indicate insertions). In panels C and D, above each gene, the incidence of unique SNV substitutions reported in the ExAC database of unrelated individuals is shown by gene location, and stacked histograms are colored by ExAC population frequency (red, 0.001% to 0.01%; purple, 0.01% to 0.1%; light purple, 0.1% to 1%; blue, 1% to 5%; teal, 5% to 20%; green, >20%); histogram bins are 100 nucleotides. For both genes, there was no annotated variation reported in the 3′UTRs after position c.*52.
As a result of our NGS approach, we sequenced both the F8 and F9 genes in all hemophilia patients, regardless of hemophilia type. In doing so, we incidentally discovered DNA variants on the “other” gene (the nondisease-associated gene), 11 of which had been previously reported in hemophilia1-4 (see Tables 4 and 5). Interestingly, all of these variants are also reported in the ExAC database, which spans 60 706 unrelated individuals sequenced as part of disease-specific (no bleeding phenotypes) and population genetic studies.23 These include 10 variants for which individuals were found to be either homozygous or hemizygous for the variant. We recommended that the HTC provider test activity of the corresponding coagulation factor in these individuals. In the case of 2 male hemophilia A patients with incidentally detected F9 variants, F9 c.19A>T (p.Ile7Phe) and F9 c.907C>T (p.His303Tyr), providers found normal FIX levels (58% and 90%, respectively), proving that these 2 F9 variants do not cause hemophilia B.
Variants detected incidentally in MLOF that had previously been reported in hemophilia: hemophilia A patients with F9 variants detected
| Hemophilia A patient and detected F8 variant . | Incidentally detected variation in the F9 gene . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient characteristics . | F8 variant . | Variant detected in the F9 gene . | Incidence in ExAC database . | ||||||
| Sex . | Baseline FVIII level (%) . | HGVS F8 cDNA . | HGVS FVIII protein . | HGVS F9 cDNA . | HGVS FIX protein . | Previously reported in hemophilia B . | Allele count/total alleles . | Number of homozygotes . | Number of hemizygotes . |
| M | <1 | c.3637delA | p.llel213Phefs*5 | c.19A>T | p.Ile7Phe | Mild, severe, unclassified, and compound with F9 (p.Arg384*)2,4 ,* | 81/87 612 | 0 | 29 |
| M | <1 | c.6429+?_6430-?inv | (F8 inversion) | ||||||
| M | 50 | c.6533G>A | p.Arg2178HiS | ||||||
| M | 5 | c.839G>T | p.Gly280Val | c.907C>T | p.His303Tyr | Mild and severe2,4 ,† | 50/85 682 | 1 | 10 |
| M | 20 | c.21490T | p.Arg717Trp | ||||||
| M | 3 | c.5822A>G | p.Asnl941Ser | c.967G>A | p.Glu323Lys | Mild2,4 | 85/87 196 | 0 | 20 |
| M | <1 | c.6429+?_6430-?inv | (F8 inversion) | c.1346G>A | p.Arg449Gln | Mild2,4 | 10/86 953 | 0 | 7 |
| Hemophilia A patient and detected F8 variant . | Incidentally detected variation in the F9 gene . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient characteristics . | F8 variant . | Variant detected in the F9 gene . | Incidence in ExAC database . | ||||||
| Sex . | Baseline FVIII level (%) . | HGVS F8 cDNA . | HGVS FVIII protein . | HGVS F9 cDNA . | HGVS FIX protein . | Previously reported in hemophilia B . | Allele count/total alleles . | Number of homozygotes . | Number of hemizygotes . |
| M | <1 | c.3637delA | p.llel213Phefs*5 | c.19A>T | p.Ile7Phe | Mild, severe, unclassified, and compound with F9 (p.Arg384*)2,4 ,* | 81/87 612 | 0 | 29 |
| M | <1 | c.6429+?_6430-?inv | (F8 inversion) | ||||||
| M | 50 | c.6533G>A | p.Arg2178HiS | ||||||
| M | 5 | c.839G>T | p.Gly280Val | c.907C>T | p.His303Tyr | Mild and severe2,4 ,† | 50/85 682 | 1 | 10 |
| M | 20 | c.21490T | p.Arg717Trp | ||||||
| M | 3 | c.5822A>G | p.Asnl941Ser | c.967G>A | p.Glu323Lys | Mild2,4 | 85/87 196 | 0 | 20 |
| M | <1 | c.6429+?_6430-?inv | (F8 inversion) | c.1346G>A | p.Arg449Gln | Mild2,4 | 10/86 953 | 0 | 7 |
FIX level available in MLOF: male FIX level 58%.
FIX level available in MLOF: male FIX level 90%.
Variants detected incidentally in MLOF that had previously been reported in hemophilia: hemophilia B patients with F8 variants detected
| Hemophilia B patients and F9 variant detected . | Incidentally detected variation in the F8 gene . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient characteristics . | Reported F9 variant . | Variant detected F8 gene . | Incidence in ExAC database . | ||||||
| Sex . | Baseline FIX level (%) . | HGVS F9 cDNA . | HGVS FIX protein . | HGVS F8 cDNA . | HGVS FVIII protein . | Previously reported in hemophilia A . | Allele count/total alleles . | Number of Homozygotes . | Number of hemizygotes . |
| M | 4 | c.688G>A | p.Gly230Arg | c.389-9C>T | NA | Mild1 | 329/86 143 | 7 | 98 |
| M | <1 | c.709C>T | p.Gln237* | ||||||
| M | <1 | c.1115T>C | p.Leu372Pro | ||||||
| M | <1 | c.968_971dup | p.Leu325Thrfs*15 | c.3169G>A | p.Glul057Lys | Mild, moderate, and severe1,3 | 58/87 425 | 1 | 19 |
| M | <1 | c.727_728delGTinsA | p.Val243llefs*2 | c.3263C>T | p.Thrl088lle | Severe3 | 22/87 381 | 0 | 1 |
| M | 2 | c.881G>A | p.Arg294Gln | c.3342G>A | p.(= ) | Mild3 | 7/87 324 | 0 | 2 |
| M | 2 | c.881G>A | p.Arg294Gln | ||||||
| M | <1 | c.1145G>A | p.Cys382Tyr | c.6374G>C | p.Ser2125Thr | Severe and unclassified3 | 2/87 725 | 0 | 0 |
| M | <1 | c.158_165delins | p.Glu53Glyfs*10 | c.6623A>G | p.Gln2208Arg | Mild1 | 1/87 743 | 0 | 1 |
| GTAAATTGGAAG | |||||||||
| M | 8 | c.1025C>T | p.Thr342Met | c.6929C>T | p.Thr2310lle | Mild3 | 3/83 951 | 0 | 2 |
| Hemophilia B patients and F9 variant detected . | Incidentally detected variation in the F8 gene . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient characteristics . | Reported F9 variant . | Variant detected F8 gene . | Incidence in ExAC database . | ||||||
| Sex . | Baseline FIX level (%) . | HGVS F9 cDNA . | HGVS FIX protein . | HGVS F8 cDNA . | HGVS FVIII protein . | Previously reported in hemophilia A . | Allele count/total alleles . | Number of Homozygotes . | Number of hemizygotes . |
| M | 4 | c.688G>A | p.Gly230Arg | c.389-9C>T | NA | Mild1 | 329/86 143 | 7 | 98 |
| M | <1 | c.709C>T | p.Gln237* | ||||||
| M | <1 | c.1115T>C | p.Leu372Pro | ||||||
| M | <1 | c.968_971dup | p.Leu325Thrfs*15 | c.3169G>A | p.Glul057Lys | Mild, moderate, and severe1,3 | 58/87 425 | 1 | 19 |
| M | <1 | c.727_728delGTinsA | p.Val243llefs*2 | c.3263C>T | p.Thrl088lle | Severe3 | 22/87 381 | 0 | 1 |
| M | 2 | c.881G>A | p.Arg294Gln | c.3342G>A | p.(= ) | Mild3 | 7/87 324 | 0 | 2 |
| M | 2 | c.881G>A | p.Arg294Gln | ||||||
| M | <1 | c.1145G>A | p.Cys382Tyr | c.6374G>C | p.Ser2125Thr | Severe and unclassified3 | 2/87 725 | 0 | 0 |
| M | <1 | c.158_165delins | p.Glu53Glyfs*10 | c.6623A>G | p.Gln2208Arg | Mild1 | 1/87 743 | 0 | 1 |
| GTAAATTGGAAG | |||||||||
| M | 8 | c.1025C>T | p.Thr342Met | c.6929C>T | p.Thr2310lle | Mild3 | 3/83 951 | 0 | 2 |
Discussion and conclusion
Through MLOF, we have developed a robust collaboration to effectively provide genotyping for a large number of patients with hemophilia A and B in the United States. In a coordinated effort with HTCs nationwide, by the end of the project we will have enrolled ∼7000 male patients with hemophilia, which represents approximately one-third of the male hemophilia A and B patients in the US MLOF, and most of these individuals have consented to participate in research, an effort that would otherwise be limited by the rarity of these diseases. Patients and families have given input into the project, and educational sessions have provided a means to help patients and providers understand the complexities inherent in genetic testing.
For this project, we designed a novel, high-throughput method for hemophilia genotyping using an MIP-targeted NGS method. This allowed us to sequence the F8 and F9 genes and screen for F8 inversions simultaneously in a large number of samples (192 or 384) at a time. Although proof-of-principle NGS has been reported for genotyping and discovery in patients with bleeding disorders, <50 hemophilia A and B patients have been previously reported using NGS in approaches that cannot detect F8 inversions.24-26
In the first 3000 MLOF patients reported here, the spectrum of types of F8 and F9 genetic variants we found was similar to that previously reported in hemophilia, including in a report of 829 US patients with hemophilia A or B.1-5,12,27-30 Surprisingly, despite a long history of genetic studies in hemophilia, we identified 273 previously unreported F8 and F9 DNA variants, significantly advancing our knowledge of the genetics of hemophilia. Unique novel variants continued to be identified as the first 3000 patients were enrolled, suggesting that more novel variants will be detected as the project continues.
DNA variants found were similar to those previously reported by other investigators and in the hemophilia databases.1-5,27-30 Variants predicted to be gene-disrupting changes were detected predominantly in males with severe disease, as expected. SVs were more common in hemophilia A because of F8 intron 22 and intron 1 inversions, which accounted for 43% of severe male cases. The incidence of other large SVs was 6% and 10% in severe male hemophilia A and B, respectively. In F8, complex intron 22 and intron 1 inversions, Alu insertions, and a complex partial exon 14 duplication were also detected. These data support the need for dedicated assessments of structural variation in the genotyping of hemophilia patients, particularly patients without a variant detected by other means.
There have been reports of patients with more than 1 likely deleterious variant,29-34 including in hemophilia B, but the incidence of multiple such variants in hemophilia patients has not been determined. In some cases, the 2 variants likely represent linked recombination events (see Tables 2 and 3). This has been previously reported in hemophilia in association with intron 22 inversions.35,36
This study offers the opportunity to discover the incidence of multiple reportable variants in hemophilia patients, as all subjects had gene sequencing and inversion testing simultaneously. We find that more than 1 reportable DNA variant was detected in ∼0.1% of patients. In these males, zygosity was easily established, as all males tested were hemizygotes, and therefore their multiple F8 or F9 variants must lie in cis. Multiple variants on the same allele could result from recombination between alleles carrying these variants singly or de novo variation arising on an allele that already harbored a variant. In females, multiple variants could lie in cis or in trans. There are significant implications in counseling a woman with 2 variants that may lie in trans, as all of her sons would be affected by hemophilia, and her daughters would be obligate carriers and possibly be affected by hemophilia. Family studies outside the scope of the MLOF study would be needed to investigate inheritance and pathogenicity of the multiple variants detected in females. In 14 males with severe disease and 2 reportable variants, at least one of the variants was a predicted null variant, making assessment of pathogenicity of the second variant impossible without additional information. However, of these second variants detected in cis with predicted null variants, 11 had been previously reported alone in other patients with hemophilia, enabling interpretation of clinical significance of both alleles in those cases.
We were unable to find a potentially causative variant in 48 hemophilia patients, 44 of whom have hemophilia A, 69% of whom had mild disease. One possibility is that genetic variants that adversely impact F8 or F9 gene function and cause hemophilia lie in genomic regions outside the target captured in our NGS and Sanger sequencing methods. For example, multiple groups have reported rare F8 deep intronic variants as the cause of hemophilia A in patients for whom a variant had not been found.37-39 It is also likely that some of the patients diagnosed with hemophilia A, particularly those with mild disease, instead have low FVIII because of undiagnosed von Willebrand disease.40 Thus, in patients diagnosed with hemophilia A where a clinically reportable variant was not identified, we recommended that the provider evaluate the patient for von Willebrand disease. In the near future, ∼5000 MLOF Research Repository samples will undergo whole genome sequencing through the US National Heart, Lung and Blood Institute Transomics in Precision Medicine program, through which we may be able to determine the genetic cause of hemophilia in patients who did not have a variant identified by this targeted sequencing method and to study other research questions in hemophilia.
Outside of the region encoding the FVIII B domain, the F8 and F9 genes have been purported to be highly conserved with little benign variation in the coding regions, and it has been clinical practice to assume that DNA variants detected in F8 or F9 are the cause of the patient’s hemophilia A or B, respectively. This assumption has been questioned for F8,41 and a recent study of F8 genetic variation in the 1000 Genomes Project further supported the presence of considerable benign variation in the F8 gene across ethnic groups.42 In MLOF, we identified numerous likely nondeleterious variants in both the F8 and F9 genes (supplemental Table 2), and normal factor levels demonstrate that at least 2 variants previously reported in hemophilia are benign variants. Interrogation of the ExAC database for F8 and F9 variants captured in whole exome sequencing of ∼61 000 individuals43 further supports that there is considerable rare variation in both genes (shown in Figure 4C-D and supplemental Table 6). Understanding that not all F8 and F9 genetic variation causes hemophilia is essential to avoid overassigning clinical significance to variants in the interpretation of clinical hemophilia genotype data. Analysis of clinically reported F8 and F9 variants compared with unreported variants showed that reported MLOF variants were significantly less likely to be present in the ExAC database (F8: odds ratio = 63.4, P < .001; F9: odds ratio = 283.5, P < .001), consistent with the expectation for enrichment of variants that impact gene function in disease cohorts relative to nondiseased populations (supplemental Table 7).
Instances of incorrect interpretation of DNA variant pathogenicity that impacted clinical care have been documented.44 To reduce this risk, new guidelines for interpretation of DNA variants have been developed, including by the American College of Medical Genetics and Genomics.22,45 Traditionally in hemophilia, only 1 affected member of a family has been genotyped. This precludes familial segregation data and genetic confirmation of suspected de novo variation that can be used to support interpreting the pathogenicity of a variant. Through the MLOF project, we are working to obtain data in families to establish segregation of variants or de novo occurrence to support classification of F8 and F9 variants as functional (pathogenic) or benign. Through these and other efforts, we should be able to strengthen genetic data in hemophilia and further inform annotation of variants in existing databases.
In addition to returning results to patients, MLOF F8 and F9 gene variant data will be shared through the EAHAD1,2 and CDC3,4 public F8 and F9 variant databases. The EAHAD databases also curate information as to the number of times variants have been reported in hemophilia; thus, all variants found, including those previously reported, are being deposited in EAHAD.1,2 We expect that additional novel F8 and F9 genetic variation will be detected as the MLOF project progresses.
This project has several limitations. Factor levels that inform hemophilia severity were reported by the HTCs and not determined centrally, which could lead to misrepresentation of baseline levels because of variance in local laboratories or other errors, such as the patient having exogenous factor (drug) circulating at the time of the blood draw. For example, a few predicted null DNA variants were detected in male patients with reported factor levels of 1% to 5%, which is likely erroneous but resulted in assignment of a moderate rather than severe hemophilia disease severity. In such circumstances, we contacted HTCs to confirm factor activity levels, but we were not able to verify the baseline levels for all such apparent discrepancies. Additionally, as described previously, our high-throughput genotyping method was designed to detect variants in the F8 and F9 open reading frames, splice sites, F8 inversions, and larger SVs impacting the coding regions. A broader approach to sequencing would have likely allowed us to detect additional DNA variants of interest in noncoding regions. Lastly, even though this is a large collaborative project, it has been limited by staff availability and patient access.
A major strength of the MLOF project is the establishment of a research repository containing DNA sequence, DNA, RNA, serum, and plasma obtained under an institutional review board–approved protocol that includes consent for whole genome sequencing. In the near future, investigators will be able to apply for access to the deidentified genetic data, repository samples, and phenotypic data through ATHN.
In conclusion, US HTCs and their patients enthusiastically embraced MLOF and enabled this genetic study of unprecedented scale in hemophilia A and B, rare diseases affecting 1:5000 live male births. Successful collaboration across partners and with the HTCs that deliver clinical and genetic services has been integral to the program’s success. Using our custom high-throughput sequencing platform we were able to detect F8 inversion variants simultaneously with other sequence variants. Actionable DNA variants were found in almost all subjects, and many novel clinically reportable hemophilia variants were found. The data from this project are advancing our understanding of hemophilia and enhancing our ability to accurately interpret the significance of F8 and F9 genetic variants in patients with hemophilia. Furthermore, the MLOF Research Repository, by providing data and samples for genotype/phenotype correlations, will support research to improve our understanding of hemophilia and its complications and to advance hemophilia care.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors wish to acknowledge all of the HTC staff who enrolled patients, the participating patients, and their families. The authors also wish to acknowledge other members of the BWNW Project Team: Sarah Heidl, Angela Dove, Kristen Koltun, Sarah Ryan, Ann Whitney, Cierra Leon-Guerrero, and Gayle Teramura.
This work was supported by Bioverativ, which provided funding to perform the genotyping at no cost to patients and to establish the MLOF repository.
Authorship
Contribution: J.M.J. and S.N.F participated in study methods design, data analysis, interpretation of results, and writing the manuscript; H.H. participated in data analysis, interpretation of results, and writing the manuscript; S. Roberge participated in sample acquisition, data analysis, and writing the manuscript; B.K.M. participated in study method design, data analysis, and writing the manuscript; M.K. participated in study design, data analysis, and writing the manuscript; N.C.J. participated in clinical study design, study methods design, data analysis, and writing the manuscript; J.S. participated in study methods design, data analysis, interpretation of results, and writing the manuscript; S. Ruuska participated in sample acquisition, data analysis, and writing the manuscript; M.A.K., J.M., D.J.A., and G.F.P. participated in study design and writing the manuscript; B.A.K. participated in study design, data analysis, interpretation of results, and writing the manuscript; and all authors had full editorial control of the content and provided their final approval before publishing.
Conflict-of-interest disclosure: J.M. is an employee of Bioverativ (which funded MLOF). The remaining authors declare no competing financial interests.
Correspondence: Barbara A. Konkle, Bloodworks Northwest, 921 Terry Ave, Seattle, WA 98104; e-mail: barbarak@bloodworksnw.org.

![Figure 4. Schematic of the locations of single nucleotide substitutions (single nucleotide variants [SNVs]), excluding nonsense variants; SVs in hemophilia A and B; and Exome Aggregation Consortium (ExAC) database SNVs relative to the F8 and F9 cDNAs. Scale schematics of the coding regions of the F8 gene (A, C) and the F9 gene (B, D). UTRs are depicted with blue filled boxes; exons are depicted as open boxes and numbered beneath. In panels A and B, above each gene, the frequency of unique SNV substitutions (missense, synonymous, splice, and UTR) by gene location is shown for males, and stacked histograms are colored by hemophilia disease severity (red, severe; purple, moderate–severe; light purple, mild–severe; blue, moderate; teal, mild–moderate; green, mild); histograms bins are 100 nucleotides. Below each gene, regions impacted by unique SVs are shown (red bars indicate deletions, blue bars indicate duplications, and pink hexagons indicate insertions). In panels C and D, above each gene, the incidence of unique SNV substitutions reported in the ExAC database of unrelated individuals is shown by gene location, and stacked histograms are colored by ExAC population frequency (red, 0.001% to 0.01%; purple, 0.01% to 0.1%; light purple, 0.1% to 1%; blue, 1% to 5%; teal, 5% to 20%; green, >20%); histogram bins are 100 nucleotides. For both genes, there was no annotated variation reported in the 3′UTRs after position c.*52.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/13/10.1182_bloodadvances.2016002923/3/m_advances002923f4.jpeg?Expires=1764961398&Signature=Vo-DET-HGhhEq5LB95hWIHQvTB9QSFrG9fOMnPI91iAjxp1cWV0NeCoxBymPr~Zd7cfv70~iR2huh0RC-C46Dj-O~MU8NR3GFqqGq2atqCdNGcGuM5sWmUVyodOrkDy6kVJUuivUa~0vpej9OhQ8UUwCNunWVSblUwL8EMGxMpirEZE0kAmZDJyDOrcfy1hgOXbHnCmnJgzq9PfK5wUy0EG02oMeTyNuu1y3kqXFiX-U38RcLLgR6YPmdlWsepQjdzxGvKYbteisvJSiHKLl~s8f1znOnyd2soBwXj4fVwcJHtCw6YdzfyPv-ufH8oNwXHEeHoYRvH~93m5wQQMUYA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)