Key Points
IL-25, IL-33, and TSLP induce distinct activation profiles in ILC2s. IL-2 further amplifies their response and induces an NK-like phenotype.
ILC2 plasticity is observed in serum-free media even when in the presence of IL-25, IL-33, and TSLP, and absence of either IL-1β or IL-12.
Abstract
Innate lymphoid cells (ILCs) represent a distinct branch of the lymphoid lineage composed of 3 major subpopulations: ILC1, ILC2, and ILC3. ILCs are mainly described as tissue-resident cells but can be detected at low levels in human blood. However, unlike mouse ILCs, there is still no consistent methodology to purify and culture these cells that enables in-depth analysis of their intrinsic biology. Here, we describe defined culture conditions for ILC2s, which allowed us to dissect the roles of interleukin 2 (IL-2), IL-25, IL-33, and thymic stromal lymphopoietin (TSLP) individually, or in combination, in modulating ILC2 phenotype and function. We show that TSLP is important for ILC2 survival, while ILC2 activation is more dependent on IL-33, especially when in combination with IL-2 or TSLP. We found that activation of ILC2s by IL-33 and TSLP dramatically upregulated their surface expression of c-Kit and downregulated expression of the canonical markers IL-7Rα and CRTH2. IL-2 further amplified ILC2 production of IL-5, IL-13, and granulocyte-macrophage colony-stimulating factor but also induced a more natural killer (NK)–like phenotype in ILC2, with upregulation of granzyme B production by these cells. Furthermore, ILC2 plasticity was observed in serum-free SFEM II media in response to IL-33, IL-25, and TSLP stimulation and independently of IL-12 and IL-1β. This is the first comprehensive report of an in vitro culture system for human ILC2s, without the use of feeder layers, which additionally evaluates the impact of IL-25, IL-33, and TSLP alone or in combination on ILC2 surface phenotype and activation status.
Introduction
Type 2 innate lymphoid cells (ILC2s) were first characterized in 2010 as an interleukin 13 (IL-13)–producing non–B/non–T innate effector cell type crucial in type 2 immune responses, such as helminth infection.1 In the intervening years, the ILC field has progressed rapidly, leading to identification of group 1 and group 3 ILCs and delineation of their developmental origin from a common lymphoid progenitor in the bone marrow.2 As knowledge of these cells increases, a potential role in a range of inflammatory pathologies3 has been proposed. In particular, ILC2s have been shown to express receptors for epithelial cytokines (ECs), or “alarmins,” such as IL-25, IL-33 and thymic stromal lymphopoietin (TSLP), and to respond to these signals by producing IL-5 and IL-13, which potentiate disease phenotypes in several models.4,5 However, the relative contribution and importance of each of these cytokines in directing the ILC phenotype are yet to be conclusively established. Studies in mouse models suggest that IL-33 may be more potent than IL-25 or TSLP in directing allergic inflammation in the lungs via ILC activation,4 while redundancy among the 3 cytokines was found with respect to their role during chronic inflammation and fibrosis.6
While most of our knowledge of ILCs and their biology has come from mouse models, ILCs have been identified in human blood, skin, lung, and gut, where their increased numbers are associated with severity in several chronic diseases, like asthma, atopic dermatitis, chronic obstructive pulmonary disease (COPD), and inflammatory bowel disease.7-9 The first description of a methodology to purify an ILC population from human peripheral blood identified CRTh2 and CD161 as markers of group 2 ILCs.8 Although progress has been made in investigating the biology of these cells in vitro, there is still a lack of a common methodology for their isolation and culture, and most described methods expand ILCs in coculture with feeder layers, including irradiated peripheral blood mononuclear cells (PBMCs), which introduce variables to the culture system. This limitation, together with recently evidenced plasticity of ILCs,10-12 can confound the ability to understand ILC intrinsic biology in vitro and make the development of a system where all variables can be easily controlled imperative to greater understanding of this increasingly important cell type.
Here, we describe a rapid, reliable, and robust method of isolating pure populations of ILC1s, ILC2s, and ILC3s from human blood using lineage depletion followed by cell sorting based on expression of CD127 (IL-7Rα) and CRTh2 and CD117 (c-Kit), but not CD161. In vitro culture of ILC2s showed that different baseline media choices can have a significant impact on survival, cell phenotype, effector function, and plasticity, even in the absence of IL-12 and IL-1β stimuli.
ECs are an attractive therapeutic target in several inflammatory diseases, and understanding the relative role that ECs play in modulating ILC behavior is critical, especially because ILCs are emerging as key players in directing other cell types, such as dendritic cells, T cells, and even B cells,13-15 in such settings. Here, we show a differential role for TSLP in cell survival but a critical role for IL-33 in effector function, particularly in combination with other ECs or IL-2. We demonstrate that expression of canonical markers such as CRTh2 and CD127 are downregulated in response to ECs, which has potential implications for detecting ILC2s in mixed culture with other hematopoietic cell types (or feeder layers). Finally, by employing high-dimensional cytometry and visualization of high dimensional single-cell data based on t-distributed stochastic neighbor embedding algorithm (viSNE) plots,16 we further define the EC receptor expression pattern and heterogeneity within ILC2 cultures. The role of IL-2 in ILC2 activation was also evaluated, and we found that IL-2, when combined with IL-33, but not individually, can further increase granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-5, and IL-13 production in culture. However, this increased ILC2 activation comes accompanied with a change in their phenotype to a more natural killer (NK)–like ILC, with upregulation of the NK receptor NKp30 as well as granzyme B (but not perforin) production. This is the first study that evaluates in depth the phenotype and activation status of human ILC2, and the relative roles of IL-25, IL-33 and TSLP, in an in vitro culture system without the presence of other feeder cells.
Methods
ILC2 isolation
PBMCs were isolated from healthy human blood (National Health Service Blood and Transplant Services) using a Sepmate column (STEMCELL Technologies) and Ficoll-Paque Plus (GE Healthcare). For ILC enrichment, EasySep NK-cell–negative selection (STEMCELL Technologies) and EasySep CD56-positive selection II (STEMCELL Technologies) were used in combination, per the manufacturer’s instructions, as this achieved the highest enrichment for the ILCs (supplemental Figure 1). Following enrichment, 106 cells/mL were stained in fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline, 10% fetal bovine serum, and 2 mM EDTA) for 30 minutes at 4°C, in the dark, using the antibody cocktail described in Table 1. Cells were sorted at a density of 5 × 106 to 107 cells/mL with a Becton Dickinson ARIAIII. Prior to cell sorting, 4′,6-diamidino-2-phenylindole (DAPI) was added to the cells at a concentration of 1/2500 to enable exclusion of dead cells.
Antibody panel for cell sorting and phenotyping by flow cytometry
| Antigen . | Fluorophore . | Clone . | Supplier . | Concentration (μL/1e6 cells) . |
|---|---|---|---|---|
| CD45 | V500 | HI30 | BD Biosciences | 5 |
| CRTH2 | Alexa Fluor 647 | BM16 | BioLegend | 5 |
| CD117 | PECy7 | 104D2 | eBioscience | 5 |
| CD127 | BV421 | HIL-7R-M21 | BD Biosciences | 5 |
| NKp44 | PE | P44-8 | BioLegend | 5 |
| Lineage panel | ||||
| CD11b | FITC | ICRF44 | BioLegend | 5 |
| CD14 | FITC | M5E2 | BD Biosciences | 0.61 |
| CD16 | FITC | abioCB16 | BD Biosciences | 5 |
| CD303a | FITC | 201A | BioLegend | 5 |
| TCRab | FITC | IP26 | BioLegend | 2.5 |
| CD19 | FITC | HIB19 | BioLegend | 0.31 |
| CD123 | FITC | 6H6 | BioLegend | 5 |
| CD34 | FITC | 581 | BioLegend | 0.55 |
| FcERI | FITC | AER-37 | BioLegend | 5 |
| CD1a | FITC | HI149 | eBioscience | 0.17 |
| CD94 | FITC | HP-3D9 | BD Biosciences | 5 |
| CD3 | FITC | UCHT1 | BioLegend | 1.67 |
| CD20 | FITC | L27 | BD Biosciences | 1.67 |
| CD11c | FITC | 3.9 | eBioscience | 5 |
| TCRgd | FITC | B1 | BioLegend | 5 |
| Brilliant stain buffer | n/a | n/a | BD Biosciences | 5 |
| Antibodies used for cell phenotyping | ||||
| CD117 | BV650 | 104D2 | BD Biosciences | 5 |
| CRTH2 | PE | BM16 | eBioscience | 5 |
| TSLPR | PerCP-efluor710 | eBio1A6 | eBioscience | 2.5 |
| IL-17BR | Unconjugated | D9.2 | n/a | 1.25 |
| T1/ST2 | FITC | B4E6 | MD Biosciences | 2.5 |
| HLA-DR | APCCy7 | G46-6 | BD Biosciences | 5 |
| Gata 3 | BV711 | L50-823 | BD Biosciences | 2.5 |
| Rorgt | BV650 | Q21-559 | BD Biosciences | 5 |
| T-bet | PECy7 | 4B10 | BioLegend | 5 |
| CD117 | PE | 104D2 | BioLegend | 5 |
| CD161 | APCCy7 | HP-3G10 | BioLegend | 5 |
| NKp30 | BV605 | p30-15 | BD Horizon | 5 |
| NKG2D | PerCpCy5.5 | 1D11 | BioLegend | 5 |
| IL-13 | BV711 | JES10-5A2 | BD Biosciences | 2.5 |
| IL-5 | PE | JES1-39D10 | BioLegend | 2.5 |
| IL-22 | PerCPefluor710 | 22URTI | eBioscience | 3 |
| IL-17A | BV605 | BL168 | BioLegend | 2.5 |
| IFN-γ | APCefluor780 | 4S.B3 | eBioscience | 3 |
| Granzyme B | FITC | GB11 | BioLegend | 2.5 |
| Perforin | APCCy7 | dG9 | BioLegend | 2.5 |
| CRTH2 | PEDazzle | BM16 | BioLegend | 5 |
| Antigen . | Fluorophore . | Clone . | Supplier . | Concentration (μL/1e6 cells) . |
|---|---|---|---|---|
| CD45 | V500 | HI30 | BD Biosciences | 5 |
| CRTH2 | Alexa Fluor 647 | BM16 | BioLegend | 5 |
| CD117 | PECy7 | 104D2 | eBioscience | 5 |
| CD127 | BV421 | HIL-7R-M21 | BD Biosciences | 5 |
| NKp44 | PE | P44-8 | BioLegend | 5 |
| Lineage panel | ||||
| CD11b | FITC | ICRF44 | BioLegend | 5 |
| CD14 | FITC | M5E2 | BD Biosciences | 0.61 |
| CD16 | FITC | abioCB16 | BD Biosciences | 5 |
| CD303a | FITC | 201A | BioLegend | 5 |
| TCRab | FITC | IP26 | BioLegend | 2.5 |
| CD19 | FITC | HIB19 | BioLegend | 0.31 |
| CD123 | FITC | 6H6 | BioLegend | 5 |
| CD34 | FITC | 581 | BioLegend | 0.55 |
| FcERI | FITC | AER-37 | BioLegend | 5 |
| CD1a | FITC | HI149 | eBioscience | 0.17 |
| CD94 | FITC | HP-3D9 | BD Biosciences | 5 |
| CD3 | FITC | UCHT1 | BioLegend | 1.67 |
| CD20 | FITC | L27 | BD Biosciences | 1.67 |
| CD11c | FITC | 3.9 | eBioscience | 5 |
| TCRgd | FITC | B1 | BioLegend | 5 |
| Brilliant stain buffer | n/a | n/a | BD Biosciences | 5 |
| Antibodies used for cell phenotyping | ||||
| CD117 | BV650 | 104D2 | BD Biosciences | 5 |
| CRTH2 | PE | BM16 | eBioscience | 5 |
| TSLPR | PerCP-efluor710 | eBio1A6 | eBioscience | 2.5 |
| IL-17BR | Unconjugated | D9.2 | n/a | 1.25 |
| T1/ST2 | FITC | B4E6 | MD Biosciences | 2.5 |
| HLA-DR | APCCy7 | G46-6 | BD Biosciences | 5 |
| Gata 3 | BV711 | L50-823 | BD Biosciences | 2.5 |
| Rorgt | BV650 | Q21-559 | BD Biosciences | 5 |
| T-bet | PECy7 | 4B10 | BioLegend | 5 |
| CD117 | PE | 104D2 | BioLegend | 5 |
| CD161 | APCCy7 | HP-3G10 | BioLegend | 5 |
| NKp30 | BV605 | p30-15 | BD Horizon | 5 |
| NKG2D | PerCpCy5.5 | 1D11 | BioLegend | 5 |
| IL-13 | BV711 | JES10-5A2 | BD Biosciences | 2.5 |
| IL-5 | PE | JES1-39D10 | BioLegend | 2.5 |
| IL-22 | PerCPefluor710 | 22URTI | eBioscience | 3 |
| IL-17A | BV605 | BL168 | BioLegend | 2.5 |
| IFN-γ | APCefluor780 | 4S.B3 | eBioscience | 3 |
| Granzyme B | FITC | GB11 | BioLegend | 2.5 |
| Perforin | APCCy7 | dG9 | BioLegend | 2.5 |
| CRTH2 | PEDazzle | BM16 | BioLegend | 5 |
FITC, fluorescein isothiocyanate; n/a, not applicable; PE, phycoerythrin.
In vitro culture
ILC2 populations with >98% purity were used for cell culture and cultured at a density of 500 cells per well in 96-well U-bottom tissue culture plates for 5 days in either Iscove modified Dulbecco medium (IMDM)/10% fetal bovine serum or Stempan SFEM II (STEMCELL Technologies), both with 1× penicillin/streptomycin. Cytokine dose responses were established for ILC2s, and in all studies, we selected 500 ng/mL as the optimal cytokine condition to distinguish among single, double, and triple cytokine combinations (Table 2). For intracellular staining experiments, cells were plated at a density of 1000 to 2000 cells per well.
Flow cytometry
Cells were centrifuged for 10 minutes at 400g, and supernatants collected and stored at −80°C. Cells were resuspended in FACS buffer with 10% human serum containing Fc block and antibody cocktail as described in Table 1. After a 30-minute incubation at 4°C in the dark, cells were washed once in FACS buffer and resuspended in FACS buffer containing DAPI. For IL-17BR staining, the antibody was labeled using the Zenon Alexa 700 labeling kit (Thermo Fisher Scientific) 5 minutes prior to staining, as per the manufacturer’s instructions. For intracellular cytokine staining, ILCs were stimulated for 3 to 4 hours with brefeldin A, with or without phorbol myristate acetate/ionomycin. Cells were then prepared for staining using the BD Fix/Perm kit according to the manufacturer’s instructions. For transcription factor analysis, the eBioscience Foxp3 kit was used. Samples were acquired using a BD Fortessa and data analyzed using FlowJo v.10. Mean fluorescence intensity was calculated based on the geometric mean of the curve. For viSNE analysis, raw fcs files were uploaded to Cytobank.17 Live events were gated based on exclusion of the DAPI dye, and viSNE analysis16 was conducted clustering on the expression of ST2, TSLPR, and IL17BR. Analysis was repeated 3 times in order to confirm the reproducibility and stability of clustering.
ELISA
Cytokine analyses were performed using a Th1/Th2 multiplex assay and U-plex IL-22 enzyme-linked immunosorbent assay (ELISA; Mesoscale) as per the manufacturer’s instructions.
Statistical analysis
One-way analysis of variance (ANOVA) with Dunnet correction for comparisons to control or Tukey (with a 95% confidence interval) when comparing between treatments was performed using GraphPad Prism v.6. Differences were considered significant if P ≤ .05.
Results
Isolation and culture of ILCs from blood
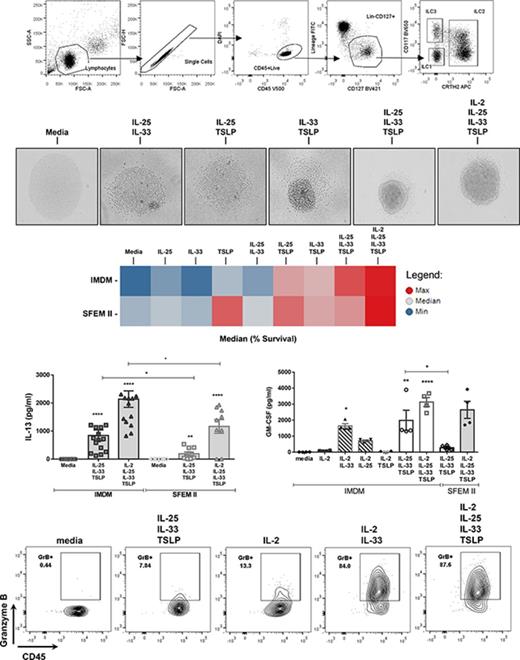
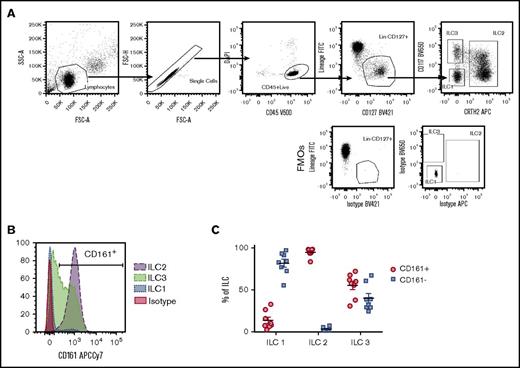
To establish a system where all ILC populations could be reliably isolated with high purity from peripheral blood, we explored a variety of lineage depletion techniques using EasySep column-free magnetic systems. CD4+ T-cell enrichment, followed by culture with IL-25, TSLP, and IL-33, was found to expand a lineage-marker–negative population that expressed high levels of IL-13 (A.C., unpublished data), but attempts to further purify ILCs from such cultures by cell sorting yielded ILC2s of inconsistent purity (supplemental Figure 1A-C). In contrast, enrichment for NK cells, which derive from the same common progenitor as ILCs,18 consistently achieved higher purity of ILC2 populations but was inadequate with respect to ILC1 and ILC3 purity, most likely due to their reduced frequency in circulation when compared with ILC2s, which are four- to 10-fold more frequent (supplemental Figure 1D). This prompted us to employ multiple enrichment steps, with the aim of maximally enriching for ILCs ahead of cell sorting. Two alternative strategies were employed, the first combining a CD4+ T-cell enrichment and CD4+ T-cell depletion (CD4 combo) and the second combining an NK-cell enrichment followed by a CD56+ NK-cell depletion (NK combo). By increasing the total ILC percentage presort from <0.1% to 4% of CD45+ cells, the latter method consistently yielded populations with a purity >98% for ILC2 and ILC3 and >95% for ILC1 following cell sorting (supplemental Figure 1E). Cells were sorted using a combination of CD45, lineage, CD127, CRTh2, and CD117 antibodies (Table 1; Figure 1A), but not CD161. We found that >98% of circulating ILC2s expressed CD161 (Figure 1B-C), making this marker redundant in isolating ILC2s. However, expression of surface CD161 in fresh circulating ILC1 and ILC3 was variable, with <20% of the ILC1 population and ∼60% of ILC3s being CD161+ (Figure 1B-C). As such, exclusion of CD161 from cell-sorting panels was able to maximize recovery of ILC1 and ILC3 populations without significantly impacting the purity or recovery of ILC2s.
Phenotype of leukocyte cone–derived ILC populations (A) Gating strategy used to purify ILC1s, ILC2s, and ILC3s from peripheral blood, in comparison with fluorescence minus one control controls, following pre-enrichment with NK combo protocol. Dot plots are representative of >10 independent experiments (n = 2 donors/experiment). (B) Differential expression of CD161 across the 3 ILC populations as depicted by a representative histogram. (C) Graph representative of the percentages of CD161+ and CD161− populations for each of the ILC types. Data are shown as mean of the population; each dot represents 1 donor. FITC, fluorescein isothiocyanate; FMO, fluorescence minus one; FSC, forward scatter; SSC, side scatter.
Phenotype of leukocyte cone–derived ILC populations (A) Gating strategy used to purify ILC1s, ILC2s, and ILC3s from peripheral blood, in comparison with fluorescence minus one control controls, following pre-enrichment with NK combo protocol. Dot plots are representative of >10 independent experiments (n = 2 donors/experiment). (B) Differential expression of CD161 across the 3 ILC populations as depicted by a representative histogram. (C) Graph representative of the percentages of CD161+ and CD161− populations for each of the ILC types. Data are shown as mean of the population; each dot represents 1 donor. FITC, fluorescein isothiocyanate; FMO, fluorescence minus one; FSC, forward scatter; SSC, side scatter.
TSLP provides a survival advantage to ILC2s
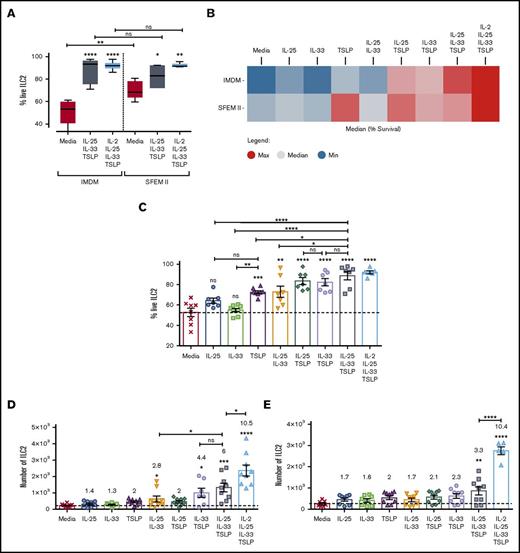
Having established a robust methodology for ILC isolation, we focused on how ECs could directly impact ILC2 behavior in vitro, and we assessed whether the redundancy that has been previously described for these cytokines6 could also applied to ILC2 activation. ILC2s were stimulated with EC alone or in combination. Alternative culture media were assessed, and SFEM II was found to provide a survival advantage, independently of cytokine stimulation, compared with IMDM (Figure 2A) or RPMI (data not shown). In media alone, 70% of cells were viable after 5 days in culture in SFEM II vs 50% in IMDM (Figure 2A). Optimal viability was achieved when cells were cultured with all 3 ECs (EC×3), or a combination of EC×3 and IL-2, regardless of baseline media (Figure 2B-C). TSLP was the only EC able to promote increased viability when provided in isolation and was additionally able to enhance the ability of IL-25 or IL-33 to do so (Figure 2B-C). In contrast, none of the ECs were able to induce proliferation on their own; however, IL-33, together with either IL-25 or TSLP, induced a population doubling within 5 days (Figure 2D). EC×3 resulted in a modest but nonsignificant further increase in proliferation of ILC2s as compared with IL-33 + TSLP stimulus, but the addition of IL-2 to EC×3 increased proliferation by 10-fold, significantly more than EC×3 alone (Figure 2D) and superior to IL-2 + IL-1β (data not shown). Choice of media was again crucial, as in SFEM II, cell proliferation was minimal unless IL-2 was present in the culture media (Figure 2E).
Survival and proliferation of ILC2s. Purified ILC2s were stimulated with different cytokine cocktails for 5 days, and their survival and proliferation were evaluated by flow cytometry. (A) Percentage of survival, as defined by exclusion of the DAPI dye was compared in cells cultured in either IMDM or SFEM II media. (B) Heat map representing the average ILC2 survival between the 2 baseline media in all stimulation cocktails, where red represent highest expression and blue lowest expression. (C) Bar graph representing each individual replicate. (D-E) ILC2 cell number, following activation with EC alone, in double or triple combination in IMDM (D) or SFEM II media (E). Numbers above each treatment group represent fold change of proliferation above media control. Data are shown as mean ± standard error of the mean (SEM) from 3 independent experiments, with n = 2/3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. ns, not significant.
Survival and proliferation of ILC2s. Purified ILC2s were stimulated with different cytokine cocktails for 5 days, and their survival and proliferation were evaluated by flow cytometry. (A) Percentage of survival, as defined by exclusion of the DAPI dye was compared in cells cultured in either IMDM or SFEM II media. (B) Heat map representing the average ILC2 survival between the 2 baseline media in all stimulation cocktails, where red represent highest expression and blue lowest expression. (C) Bar graph representing each individual replicate. (D-E) ILC2 cell number, following activation with EC alone, in double or triple combination in IMDM (D) or SFEM II media (E). Numbers above each treatment group represent fold change of proliferation above media control. Data are shown as mean ± standard error of the mean (SEM) from 3 independent experiments, with n = 2/3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. ns, not significant.
Modulation of ILC2 canonical markers is influenced by their activation status
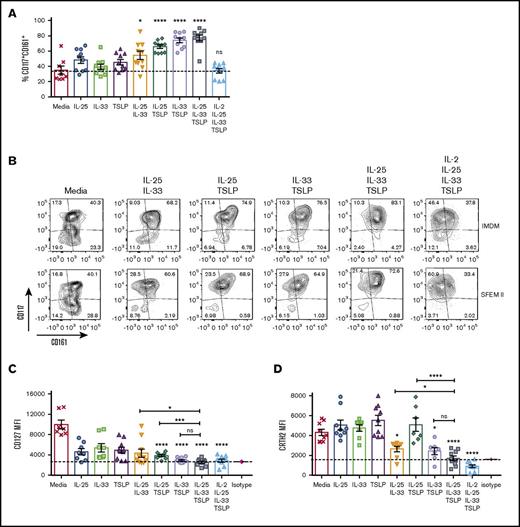
Human ILC2s are defined by surface expression of CD127, CRTH2, and CD117 and absence of lineage markers. We sought to evaluate if expression of these defining markers was modulated by EC stimulation. Freshly isolated ILC2s can be subdivided into 2 populations: CD117− (40% to 50%) and CD117+ (50% to 60%) (data not shown). This ratio was unchanged by culture in the absence of cytokines; however, the percentage of CD117+ cells increased significantly with double or triple EC stimulation (Figure 3A-B), most notably when one of the ECs was TSLP. The CD161 surface marker remained on the cell surface throughout the 5-day culture in the presence or absence of ECs but was downregulated in the presence of IL-2 (Figure 3B). CD127 is a canonical marker for ILCs; however, when ILC2s cultured in IMDM were activated in vitro, with double or triple EC combinations, CD127 surface expression was downregulated to background levels (Figure 3C). Interestingly, this downregulation was not noted when cells were cultured in SFEM II media, where expression of CD127 was low but maintained throughout the culture, regardless of stimulation (supplemental Figure 2). CRTh2 was similarly downregulated following treatment with EC combinations. IL-33 was critical to CRTh2 downregulation; however, in SFEM II media, the further addition of IL-2 was required (Figure 3D; supplemental Figure 2). Downregulation of CRTh2 expression has been reported previously in response to prostaglandin D2 (PGD2) activation of ILC219 and following migration to the lung in mice,20 but not through direct activation by ECs.
ECs differentially modulate the expression of ILC2 canonical markers. ILC2s were stimulated in vitro with ECs or EC combinations, and surface expression of CD127, CRTH2, CD161, and CD117 was evaluated by flow cytometry. (A) Percentage of CD117+CD161+ in IMDM. (B) Contour plots showing the expression pattern of CD161 (x-axis) and CD117 (y-axis) after stimulation with double or triple cytokine combinations. (C-D) Mean fluorescence intensity (MFI) was also calculated for CD127 (C) and CRTH2 (D) in IMDM. Data are shown as mean ± SEM from 2 independent experiments, with n = 2 or 3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. ns, not significant.
ECs differentially modulate the expression of ILC2 canonical markers. ILC2s were stimulated in vitro with ECs or EC combinations, and surface expression of CD127, CRTH2, CD161, and CD117 was evaluated by flow cytometry. (A) Percentage of CD117+CD161+ in IMDM. (B) Contour plots showing the expression pattern of CD161 (x-axis) and CD117 (y-axis) after stimulation with double or triple cytokine combinations. (C-D) Mean fluorescence intensity (MFI) was also calculated for CD127 (C) and CRTH2 (D) in IMDM. Data are shown as mean ± SEM from 2 independent experiments, with n = 2 or 3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. ns, not significant.
Differential activation of ILC2s by IL-2 and ECs
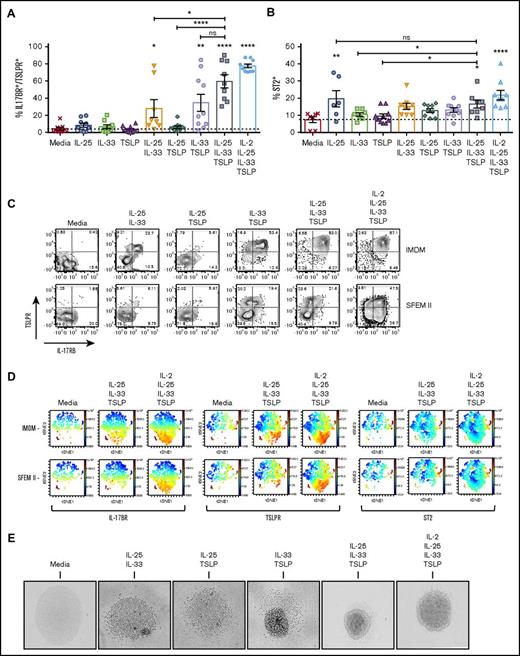
Stimulation of ILC2s induced the expression of the EC receptors TSLPR, IL17-BR, and ST2. In media alone, only 10% of ILC2s were positive for ST2 expression, 20% were positive for IL-17BR, and almost none were positive for TSLPR (Figure 4A-C). IL-25 alone induced an increase in the percentage of ST2+ cells in both IMDM and SFEM II (Figure 4B; supplemental Figure 2), but this did not significantly increase with addition of other cytokines. Moreover, the percentage of ST2+ ILC2s remained <40%, potentially due to the sensitivity of the antibody used. In contrast, when cultured in IMDM, ILC2s treated with double EC cocktails containing IL-33 demonstrated an increase in the percentage of IL-17BR and TSLPR double-positive cells from <5% to 40% to 60% and then a further increase to >80% when treated with EC×3 (Figure 4A-C). In SFEM II, ILC2s only significantly expanded this double receptor–positive population when stimulated with EC×3 and instead expanded a single-positive TSLPR or IL17BR subpopulation (Figure 4C; supplemental Figure 2).
Surface expression of TSLPR, IL-17BR, and ST2 reveals different ILC2 subpopulations. ILC2s were stimulated with ECs or EC combinations, and surface expression of EC receptors was evaluated by flow cytometry. (A) Percentage of IMDM-cultured ILC2s that show double-positive expression for IL-17BR and TSLPR. (B) Percentage of IMDM-cultured ST2+ ILC2s. (C) Contour plots showing the percentage of cells in each gate for IL-17BR (x-axis) and TSLPR (y-axis), comparing IMDM and SFEM II. (D) viSNE plots, as analyzed in Cytobank, showing the population clustering for each receptor. Red indicates the highest expression and blue the lowest expression. (E) Photographs depicting ILC2s when stimulated with double or triple cytokine combinations, or the latter combination with the addition of IL-2 (phase contrast microscopy, original magnification ×100). Data are shown as mean ± SEM from 2 independent experiments, with n = 2 or 3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above the bars represent comparison with media baseline; comparison between samples is represented by the connecting line. ns, not significant.
Surface expression of TSLPR, IL-17BR, and ST2 reveals different ILC2 subpopulations. ILC2s were stimulated with ECs or EC combinations, and surface expression of EC receptors was evaluated by flow cytometry. (A) Percentage of IMDM-cultured ILC2s that show double-positive expression for IL-17BR and TSLPR. (B) Percentage of IMDM-cultured ST2+ ILC2s. (C) Contour plots showing the percentage of cells in each gate for IL-17BR (x-axis) and TSLPR (y-axis), comparing IMDM and SFEM II. (D) viSNE plots, as analyzed in Cytobank, showing the population clustering for each receptor. Red indicates the highest expression and blue the lowest expression. (E) Photographs depicting ILC2s when stimulated with double or triple cytokine combinations, or the latter combination with the addition of IL-2 (phase contrast microscopy, original magnification ×100). Data are shown as mean ± SEM from 2 independent experiments, with n = 2 or 3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above the bars represent comparison with media baseline; comparison between samples is represented by the connecting line. ns, not significant.
Traditional flow cytometry analysis is limited in its ability to assess coexpression of >2 receptors at any one time. In order to better define the relationship between expression of TSLPR, ST2, and IL17BR in resting and activated ILC2s, the viSNE algorithm16 was used to cluster data from untreated cells and those treated with EC×3 ± IL-2 (Figure 4D). In untreated ILC2s, expression of all 3 receptors was minimal, with minor populations of ST2High/TSLPR+ double- and ST2low/TSLPR+/IL17BR+ triple-positive cells present in both IMDM and SFEM II media. Treatment with EC×3 resulted in expansion of a predominantly TSLPR+/IL17BR+ double-positive, but ST2-negative, population of cells in IMDM and, to a much lesser degree, in SFEM II. Addition of IL-2 to the triple cytokine cocktail resulted in greater expansion of this double-positive population, as well as emergence of more notable TSLPR+ or IL17BR+ single-positive populations, with the effect being most striking in SFEM II media. Increases in ST2 expression were minimal and limited predominantly to populations that expressed ST2 prior to treatment.
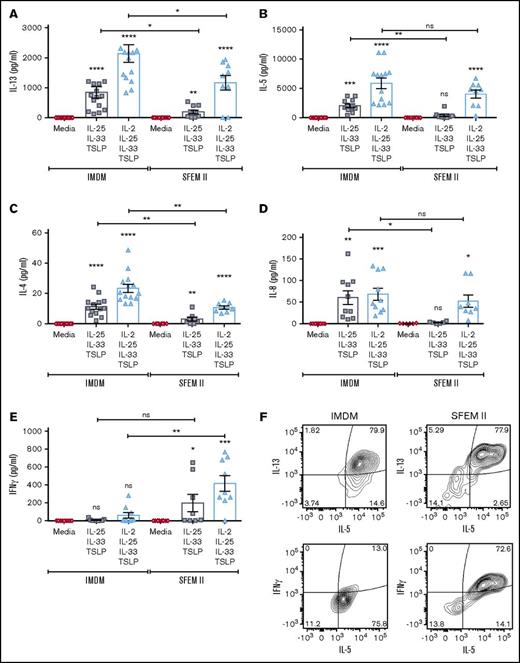
Others have found that IL-33 alone can induce high IL-5 and IL-13 production by skin-derived ILC2s in comparison with IL-25 or TSLP.19 Here, we found very little effect of the ECs by themselves on ILC2 activation, perhaps because circulating ILCs behave differently than those resident in tissue. Assessment of cytokine production in response to a dose titration of ECs only detected IL-13 or IL-5 production by ILC2s when at least 2 ECs were used (supplemental Figure 3). IL-33 was essential to ILC2 activation, and no cytokine production was measured after stimulation with IL-25 and TSLP, although high amounts of IL-13 (up to 2 ng/mL) and IL-5 (6-7 ng/mL) were secreted following stimulation with EC×3, with or without IL-2, greater than those observed following treatment with IL-2 and IL-1β (Figure 5A-B; supplemental Figure 3). Other cytokines, including IL-4 and IL-8, were also produced by activated ILC2s under these conditions but at lower levels than IL-5 or IL-13 (Figure 5C-D).
ILC2 cytokine secretion pattern following stimulation with different combinations of ECs in IMDM or SFEM II culture media. ILC2s were stimulated with the triple EC combination or with all 3 ECs plus IL-2, and effector cytokine production was quantified in the supernatants after 5 days in culture with either IMDM or SFEM II. (A-E) Multiplexed ELISA was performed to measure IL-13 (A), IL-5 (B), IL-4 (C), IL-8 (D), and IFN-γ (E). Data are shown as mean ± SEM from 2 independent experiments, with n = 2 or 3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. (F) Representative contour plots showing intracellular expression of IL-13, IL-5, and IFN-γ in ILC2s cultured in either IMDM or SFEM II following 4-hour incubation with brefeldin A. Data are representative of 2 independent experiments (n = 2 donors). ns, not significant.
ILC2 cytokine secretion pattern following stimulation with different combinations of ECs in IMDM or SFEM II culture media. ILC2s were stimulated with the triple EC combination or with all 3 ECs plus IL-2, and effector cytokine production was quantified in the supernatants after 5 days in culture with either IMDM or SFEM II. (A-E) Multiplexed ELISA was performed to measure IL-13 (A), IL-5 (B), IL-4 (C), IL-8 (D), and IFN-γ (E). Data are shown as mean ± SEM from 2 independent experiments, with n = 2 or 3 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. (F) Representative contour plots showing intracellular expression of IL-13, IL-5, and IFN-γ in ILC2s cultured in either IMDM or SFEM II following 4-hour incubation with brefeldin A. Data are representative of 2 independent experiments (n = 2 donors). ns, not significant.
Unexpectedly, ILC2s cultured in SFEM II produced significantly reduced levels of IL-13, IL-5, and IL-4; instead, we observed an upregulation of interferon γ (IFN-γ) production (Figure 5A-F). To further evaluate whether this was a true phenotypic “switch,” or a different population expanding in the culture, we analyzed cytokine production intracellularly after incubation with brefeldin A for 4 hours. We found that ILC2s stimulated with EC×3 in SFEM II could coexpress IL-5, IL-13, and IFN-γ (Figure 5F), pointing to a greater plasticity of ILC2s in this media. We also measured a significant increase in IL-17A production in the supernatants of ILC2s in SFEM II following activation by EC×3 (data not shown).
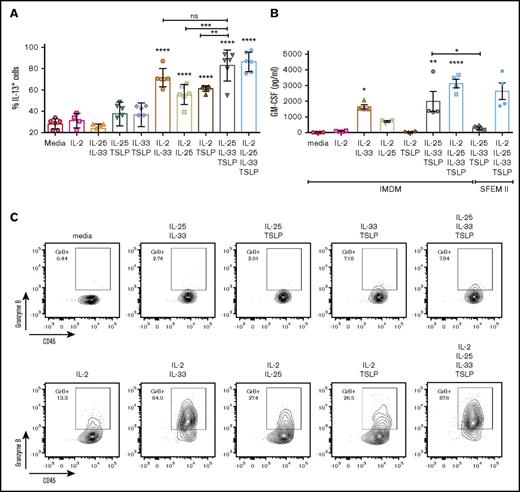
IL-2 alone, as previously reported,21 was not able to induce significant activation of ILC2s (Figure 6A-B); however, when combined with another EC (particularly IL-33), it amplified the ILC2 response, leading to an increase in IL-13 and GM-CSF production higher than EC×2 combinations. Along with these observations, we also found an increase in the cell’s granularity, which was attributed to an increased intracellular expression of granzyme B (but not perforin), when ILC2s were cultured with EC and IL-2 (Figure 6C; data not shown). In SFEM II, >90% of ILC2s expressed granzyme B intracellularly when stimulated with EC×3, even in the absence of IL-2 (data not shown). NKp30 and NKG2D surface expression were also evaluated, but we found <20% of ILC2s expressed NKp30 after EC×3 activation, and no NKG2D expression was found, independently of IMDM of SFEM II culture. We did find, however, that stimulation of ILC2s with IL-2 + IL-1β induced an upregulation of NKp30 to ∼70% to 80% of the cells and that in these conditions, 10% to 30% of ILC2s also coexpressed NKG2D (data not shown).
IL-2 amplifies the response of ILC2s to ECs. ILC2s were cultured for 5 days in the presence of EC ± IL-2. (A) Percentage of ILC2s expressing intracellular IL-13 following activation with EC ± IL-2 for 5 days and 4-hour incubation with brefeldin A prior to cell staining. Data depict 3 independent experiments (n = 2 donors/experiment). (B) GM-CSF production was also analyzed in the cells supernatants following a 5-day in vitro culture. Data depict 2 independent experiments (n = 2 donors). Data are shown as mean ± SEM from 2 independent experiments, with n = 2 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. (C) Representative contour plots of granzyme B expression by ILC2s cultured with EC ± IL-2 for 5 days and incubation with brefeldin A for 4 hours prior to intracellular cell staining. ns, not significant.
IL-2 amplifies the response of ILC2s to ECs. ILC2s were cultured for 5 days in the presence of EC ± IL-2. (A) Percentage of ILC2s expressing intracellular IL-13 following activation with EC ± IL-2 for 5 days and 4-hour incubation with brefeldin A prior to cell staining. Data depict 3 independent experiments (n = 2 donors/experiment). (B) GM-CSF production was also analyzed in the cells supernatants following a 5-day in vitro culture. Data depict 2 independent experiments (n = 2 donors). Data are shown as mean ± SEM from 2 independent experiments, with n = 2 donors each. Significance was calculated using 1-way ANOVA followed by correction for multiple comparisons, where *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistics above bars represent comparison with media baseline; comparison between samples is represented by the connecting line. (C) Representative contour plots of granzyme B expression by ILC2s cultured with EC ± IL-2 for 5 days and incubation with brefeldin A for 4 hours prior to intracellular cell staining. ns, not significant.
Discussion
TSLP, IL-25, and IL-33 have all been independently described as activators of type 2 immunity, particularly in ILC2s.22-25 However, their potency as standalone stimulators of ILC2 or their potential redundancy with each other is less clear. A better understanding of the potency of ECs as immune activators could be critical when selecting the best, most efficacious therapeutic target for diseases like asthma and COPD.
In this study, we developed a reliable methodology to quickly isolate ILC2s from peripheral blood without compromising ILC1 and ILC3 purity. For this purpose, we used an enrichment strategy combining positive and negative selection kits for NK cells using EasySep technology. We enriched for ILCs >40-fold, prior to cell sorting, and reliably isolated ILC1, ILC2, and ILC3 with >95% to 98% purity following sorting in less than 3 hours. While optimizing the sorting strategy, we found that the CD161 marker was not required for ILC2 isolation when using CD127 and CRTh2 and that if used to purify ILC1s or ILC3s, it would exclude a proportion of these cells that were CD161 negative.
We optimized culture conditions to allow for investigation of individual cytokine effects in ILC2 activation and phenotype. We found that SFEM II was optimal for viability of ILC2s, without the addition of further cytokine stimulation; however, in this media, ILC2s demonstrated several functional differences vs culture in IMDM. Their expansion and production of IL-5 and IL-13 production was minimal in the absence of IL-2, and they showed a limited ability to upregulate the receptors for TSLP and IL-25 in response to stimulus. Moreover, they appeared to show a more plastic phenotype, evidenced by coexpression of IL-5, IL-13, and IFN-γ, even in the presence of EC×3 stimulation, production of detectable levels of IL-17A in the culture supernatants (data not shown), and a trend toward an increased expression of c-Kit, a stem cell pluripotency marker.26 Since IL-12 and IL-1β have independently been shown to induce plasticity of ILC2s,10,11,21 we attempted to measure these cytokines in SFEM II but could not detect any (Table 3), suggesting that some degree of plasticity is maintained by ILC2s in this media through other unknown mediators. Another possible explanation is that ILC2s could be circulating in a less mature, progenitor-like state and that they are maintained in that developmental stage in SFEM II.
Cytokines used for in vitro culture of ILC1, ILC2, and ILC3
| Cytokine . | Supplier . | Catalog number . | Concentration (ng/mL) . |
|---|---|---|---|
| Recombinant human IL-2 | R & D Systems | 202-IL-050 | 500 |
| Recombinant human IL-23 | R & D Systems | 1290-IL-010 | 50 |
| Recombinant human IL-12 | R & D Systems | 219-IL-025 | 50 |
| Recombinant human IL-33 | MedImmune | N/A | 500 |
| Recombinant human IL-17E | Peprotech | AF-200-24 | 500 |
| Recombinant human IL-1 β | R & D Systems | 201-LB-025 | 500 |
| Recombinant human IL-7 | R & D Systems | 207-IL-005 | 10 |
| Recombinant human TSLP | R & D Systems | 1398-TS-010 | 500 |
| Cytokine . | Supplier . | Catalog number . | Concentration (ng/mL) . |
|---|---|---|---|
| Recombinant human IL-2 | R & D Systems | 202-IL-050 | 500 |
| Recombinant human IL-23 | R & D Systems | 1290-IL-010 | 50 |
| Recombinant human IL-12 | R & D Systems | 219-IL-025 | 50 |
| Recombinant human IL-33 | MedImmune | N/A | 500 |
| Recombinant human IL-17E | Peprotech | AF-200-24 | 500 |
| Recombinant human IL-1 β | R & D Systems | 201-LB-025 | 500 |
| Recombinant human IL-7 | R & D Systems | 207-IL-005 | 10 |
| Recombinant human TSLP | R & D Systems | 1398-TS-010 | 500 |
List of analytes measured in SFEM II baseline media
| Analytes undetected . | Analytes detected . | Concentration (pg/mL) . |
|---|---|---|
| GM-CSF, IL-9 | GRO-α | Below LLOD |
| IL-1β, IFN-α | EGF | Below LLOD |
| IL-1α, IL-15 | FGF-2 | Below LLOD |
| IL-12p70, IL-31 | IFN-γ | Below LLOD |
| IL-2, IL-7 | IL-8 | Below LLOD |
| Il-4, TNF-β | IL-18 | Below LLOD |
| IL-5, SCF | IL-13 | Below LLOD |
| TNF-α, IP-10 | IL-1RA | 75 ± 58 |
| IL-17A, PDGF-BB | LIF | 19 ± 2 |
| IL-21, IL-6 | HGF | 80 ± 25 |
| IL-22, IL-10 | NGF | 28 ± 2 |
| IL-23, MIP-1α | VEGF-A | 9.7 ± 1.4 |
| IL-27, MIP-1β | VEGF-D | 36 ± 3 |
| SDF-1α, MCP-1 | — | — |
| Eotaxin, RANTES | — | — |
| Analytes undetected . | Analytes detected . | Concentration (pg/mL) . |
|---|---|---|
| GM-CSF, IL-9 | GRO-α | Below LLOD |
| IL-1β, IFN-α | EGF | Below LLOD |
| IL-1α, IL-15 | FGF-2 | Below LLOD |
| IL-12p70, IL-31 | IFN-γ | Below LLOD |
| IL-2, IL-7 | IL-8 | Below LLOD |
| Il-4, TNF-β | IL-18 | Below LLOD |
| IL-5, SCF | IL-13 | Below LLOD |
| TNF-α, IP-10 | IL-1RA | 75 ± 58 |
| IL-17A, PDGF-BB | LIF | 19 ± 2 |
| IL-21, IL-6 | HGF | 80 ± 25 |
| IL-22, IL-10 | NGF | 28 ± 2 |
| IL-23, MIP-1α | VEGF-A | 9.7 ± 1.4 |
| IL-27, MIP-1β | VEGF-D | 36 ± 3 |
| SDF-1α, MCP-1 | — | — |
| Eotaxin, RANTES | — | — |
EGF, epidermal growth factor; FGF, fibroblast growth factor; GRO-α, CXCL1 or chemokine C-X-C motif ligand; HGF, hepatocyte growth factor; LIF, leukemia inhibitory factor; LLOD, lower limit of detection; MCP-1, CCL2 (monocyte chemotatic protein 1); MIP-1α, CCL3 or chemokine (C-C motif) ligand; MIP-1β, CCL4 or chemokine (C-C motif) ligand; NGF, nerve growth factor; RANTES, regulated on activation, normal T cells expressed or secreted (or CCL5); SDF, stromal-cell–derived factor; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
IL-2 has been shown in a mouse lung inflammation model to be critical for ILC2 proliferation and to synergize with IL-33 for ILC2-derived IL-13 expression.27 Here, we confirm the ability of IL-2 to amplify the response of ILC2s to IL-33, as well as IL-25 and TSLP, and additionally show a clear switch to a more NK-like phenotype, with increased granzyme B (but not perforin) production, particularly in the case of the IL-33/IL-2 combination. IL-2 + IL-1β stimulation had the same effect on granzyme B production by ILC2s, but unlike IL-33/IL-2, it was accompanied by a significant upregulation of other NK receptors such as NKp30 and NKG2D (data not shown). It is possible that IL-33, being an IL-1 family member, could play a role similar to IL-1β in inducing ILC2 plasticity in the presence of IL-2, although this would need to be further explored in future studies. Granzyme B levels have been recently correlated with the degree of pruritus and dermatitis in atopic dermatitis and psoriasis patients,28 and one could speculate that this could be coming from the increased levels of activated ILC2s.
We also found, for the first time, that IL-2 can directly downregulate the surface expression of CD161 in ILC2s. CD161 has been attributed inhibitory downstream signaling in NK cells.29,30 While the function of CD161 on ILC2s has not been fully defined, it is possible that IL-2 augments its own ability to provide activating signals to ILC2s by downregulating this potentially inhibitory receptor. In future studies, it would be interesting to understand how the IL-2–CD161 axis is modulated in ILCs and the implication of this regulation on their function, particularly in a disease setting.
There is much compelling evidence that TSLP, IL-25, and IL-33 play a role in allergic responses by acting directly on a variety of immune cell types such as T cells, dendritic cells, macrophages, and ILC2s.31,32 However, understanding their relative importance or potency has been difficult. One of the reasons for this is that high-sensitivity assays that can reliably detect these cytokines are currently not available. The first study that evaluated the combinatorial targeting of all 3 cytokines in vivo identified a redundant role for the 3 ECs and showed that type 2–driven fibrosis and inflammation could only be suppressed by disruption of all 3 signals.6 More recently, a different strategy of targeting both IL-33 and IL-13 proved superior to targeting either pathways alone in a mouse model of allergic inflammation.33 In our study, we found that ECs alone are not able to induce any notable phenotypic changes or effector functions in human ILC2s; however, a combination of IL-33 and TSLP was efficient at inducing both IL-5 and IL-13 production and significant upregulation of IL-17BR and TSLPR, which further increased when all 3 cytokines were combined, even in the absence of any other confounding aspects such as the addition of other cytokines, like IL-2 or IL-7, or the presence of other cell types. This supports the hypothesis that therapies targeting multiple pathways will be more successful at ameliorating chronic diseases like asthma and COPD. IL-33/TSLP stimulation was more potent than IL-25/TSLP or IL-25/IL-33 at inducing IL-13 and IL-5 by ILC2s, albeit at lower levels than all 3 ECs. In terms of upregulation of the receptors, IL-33 was critical at inducing an upregulation of IL-17BR and TSLPR, but the changes in ST2 expression were minimal. ST2 expression has been described as hard to detect in ILC2,34 and it is possible that better reagents will, in the future, provide better insight into its surface expression and modulation by stimuli. ILC2 responses to IL-25 were shown to be slower and less potent than IL-334 in mouse models of allergic asthma, and our results here are in keeping with IL-33 providing a stronger signal to human ILC2s in vitro. Interestingly, clustering of cells using the viSNE algorithm revealed that there are subpopulations of ILC2s that differently upregulate EC receptors following activation, highlighting the heterogeneity and multipotent character of this ILC subset. A subpopulation of ST2-positive cells clustered with a TSLPR+, but not IL17BR+, phenotype, providing some hints that although all 3 ECs can activate ILC2s, there may be a time-dependent shift in their ability to respond more strongly to a particular cytokine. In keeping with this concept, temporal regulation of IL-25 and IL-33 has been described in a mouse model of Alternaria alternata challenge, where a delayed but robust IL-25–dependent response was seen compared with IL-33.35
The expression of the stem cell factor (SCF) receptor c-Kit (CD117) and its function in ILC2s and ILC3s has seen limited exploration. Here, we found that ILC2s significantly upregulated c-Kit upon cell activation, particularly by IL-33 + TSLP or EC×3. Increased circulating SCF has been reported in both chronic asthma and pulmonary fibrosis,36,37 and SCF+ fibroblasts have been shown to induce survival and activation of c-Kit+ mast cells, a mechanism that aggravated fibrosis progression.38 It is possible that epithelial cells or fibroblasts in a disease setting could use SCF as a chemoattractant for both mast cells and ILC2s to the site of local inflammation. Indeed we have found that human ILC2s can migrate more efficiently to an SCF gradient than IL-33 in vitro (data not shown). One could hypothesize that EC×3 could upregulate c-Kit expression on ILC2s to aid their migration to the site of inflammation.
Lastly, activation of ILC2s by ECs completely downregulated their canonical markers CD127 and CRTh2. For CRTh2, this observation was previously registered in 2 independent studies in either skin-derived ILC2s following activation with PDG219 or nasal polyp–derived ILC2s following IL-2, IL-33, and TSLP stimulation. This occurred independently of a phenotypic change, as ILC2s were still producing high levels of IL-5, but not IFN-γ.11 Modulation of CD127 has been reported for T cells, and its expression can be downregulated or recycled to the surface by exposure to IL-7, IL-2, or T-cell receptor signaling.39 This study is the first report of a similar modulation of surface CD127 by human ILC2s. We found that IL33/TSLP or EC×3 stimulation completely downregulated surface expression of CD127. We observed the same finding not only in cultures of human CD4+ T cells stimulated with EC×3 but also in mouse ILC2s that had been exposed to IL-33 (data not shown). This has important implications for the understanding of ILC2 biology within mixed cultures that contain other CD45+ cells, such as irradiated PBMCs, since using flow cytometric assessment of surface markers that change in response to treatment, in particular CRTh2 and CD127, could lead to misidentification of, or failure to identify, ILC populations in such a context.
This study provides the first comprehensive description of the effects of ECs, both alone and in combination, on an in vitro culture of primary human ILC2s without the presence of feeder layers or initial cell expansion. The methodology described provides a reliable and robust in vitro model to further study ILC biology. In future studies, it will be interesting to understand how ECs can modulate other aspects of ILC2 biology, for example how they affect expression of other functional receptors like NK receptors and chemokine receptors, and to compare how tissue resident ILC2s differ from circulating cells. Lastly, this study provides further evidence that therapeutic strategies targeting multiple cytokines may be more successful than single-cytokine targeting in type 2–driven inflammatory diseases.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Andrew McKenzie for critical reading of this manuscript and Madhu Ramaswamy and Hilary Sandig for technical input.
Authorship
Contribution: A.C. and Y.O. performed all experimental work; A.C. wrote the manuscript; G.R. performed the ILC cell sorts; R.A.S. performed the Cytobank analysis and manuscript proofreading; C.O.-S. performed SFEM II–only media ELISAs; and R.D.M. and M.A.S. oversaw the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ana Camelo, MedImmune, Milstein Building, Granta Park, Cambridge CB21 0QH, United Kingdom; e-mail: cameloa@medimmune.com.