TO THE EDITOR:
Chronic lymphocytic leukemia (CLL) has high biological and clinical heterogeneity.1,2 A few prognostic factors are used in clinical practice, including immunoglobulin heavy-chain variable (IGHV) gene somatic hypermutation (SHM) status, chromosome aberrations, and gene mutations, which remain insufficient for personalized patient management.3,4 Recent studies have shown that expression of the immunoglobulin lambda light chain IGLV3-21 gene carrying an SHM-derived G>C mutation changing the glycine at position 110 to an arginine (IGLV3-21R110) defines a subset of CLL with an intermediate epigenetic profile and an aggressive clinical course.5,6 When occurring on the IGLV3-21∗01 or ∗04 alleles, the R110 mutation allows homotypic B-cell receptor (BCR) interactions, triggering cell-autonomous BCR signaling5,7 and/or facilitating T-cell–independent engagement with superantigen.8 IGLV3-21R110 has been detected in up to 6.5% of patients with CLL at diagnosis and in up to 25% of patients enrolled in clinical trials.5,6,9 We6 and others5 have shown that all CLL cases belonging to aggressive stereotyped subset #2 carried the IGLV3-21R110. Nonetheless, approximately half of IGLV3-21R110 CLL are not classified as stereotyped subset #2 but seem to have a similar clinical outcome,5,6 suggesting that the conventional stereotyped subset #2 classification might not completely recognize this clinically aggressive subgroup of CLL. In addition, IGLV3-21R110 seems to have a prognostic value independent of the IGHV gene SHM status and methylation–based epigenetic subtypes.5,6 However, further studies in independent cohorts are needed to support its application in clinical practice.1,2,10-12 The aim of this study was to assess the prognostic value of IGLV3-21R110 in large and independent population-based cohorts of patients with CLL.
We designed a multiplex IGLV3-21R110–specific polymerase chain reaction (msPCR) integrating 2 forward primers aligned to distinct regions of the IGLV3-21 gene and 2 R110-specific reverse primers matching IGLJ1 and IGLJ2/3 genes. We used a third pair of primers targeting FBXW7 as an internal control. To make the results comparable among the samples, the intensity of each IGLV3-21R110 product was normalized to the intensity of the FBXW7 band (Figure 1A; supplemental Table 1; supplemental Methods). PCR conditions were set up in a cohort of 12 patients (including 6 IGLV3-21R110 mutated) and validated in 165 CLL (including 7 IGLV3-21R110 mutated) from our previous study, in which IGLV3-21R110 status was determined using whole-genome/exome sequencing and RNA sequencing.6 The concordance was 100% (supplemental Table 2). The msPCR assay was used to characterize 2 independent CLL cohorts of 622 (cohort 1) and 376 (cohort 2) patients. To further validate the msPCR results, all but 1 IGLV3-21R110 positive sample from cohort 1 were subjected to next-generation sequencing and analyzed using IgCaller.13 The results obtained in 78 patients from cohort 2 were compared with the IGLV3-21R110 status previously reported by whole-genome sequencing and RNA sequencing.6 Fully concordant results were obtained in both validations (Figure 1B; supplemental Figures 1-2; supplemental Tables 3-9; supplemental Methods). Cohort 1 included 110 patients with high-count monoclonal B-cell lymphocytosis (MBL) and 512 with overt CLL, whereas cohort 2 included only patients with overt CLL. Although CLL patients in cohort 1 showed a shorter time to first treatment (TTFT) than those in cohort 2, no differences were observed in terms of disease stages, IGHV gene SHM status, and epigenetic subtypes (supplemental Figure 3; supplemental Table 3). The primary end point of the study was TTFT measured from the time of diagnosis, considering only the patients diagnosed with CLL (supplemental Methods). This study was approved by the Hospital Clínic of Barcelona Ethics Committee. Informed consent was obtained from all the patients.
In cohort 1, the IGLV3-21R110 mutation was found in 21 of 622 (3.4%) tumors, similarly distributed in MBL (3/110, 2.7%) and CLL (18/512, 3.5%), as well as in CLL with mutated (M-CLL) and unmutated (U-CLL) IGHV genes. Similar results were observed in cohort 2, although with a higher incidence of IGLV3-21R110 mutation (29/376, 7.7%) (Figure 1C; supplemental Table 10). Concordant with previous observations,6 IGLV3-21R110 was remarkably enriched in the epigenetic intermediate CLL (i-CLL) subtype, representing 25.8% and 41.5% of patients with i-CLL in cohorts 1 and 2, respectively (Figure 1C). IGLV3-21R110 CLL carried a borderline IGHV identity, whereas most i-CLL lacking IGLV3-21R110 had an IGHV identity below 97% (Figure 1D; supplemental Table 11). Moreover, 13 CLL with IGLV3-21R110 carried stereotyped IGH genes; 12 of 13 were classified as subset #2 (8 in cohort 1; 4 in cohort 2) and 1 as subset #14. The remaining IGLV3-21R110 CLL carried nonstereotyped immunoglobulin genes (19 major stereotyped subsets analyzed, supplemental Methods). Genomic data regarding CLL driver alterations were available for patients belonging to cohort 2. As previously reported,6 mutations in SF3B1 and ATM were more frequently found in IGLV3-21R110 CLL than in non-IGLV3-21R110 (SF3B1: 41.4% vs 10.3%; P < .0001; ATM: 25.0% vs 6.3%; P = .003). In this cohort, NOTCH1 mutations were also enriched in IGLV3-21R110 CLL (33.3% vs 10.0%; P = .003). In contrast, the IGLV3-21R110 mutation was mutually exclusive in the presence of trisomy 12 or del(17p) (trisomy 12: 0% vs 14.3%; P = .03; del(17p): 0% vs 13.1%; P = .056), as recently suggested.9 No differences were found in the distribution of TP53, BIRC3, and NFKBIE mutations, del(13q), and del(11q) (supplemental Figure 4). Clinical analyses performed in each cohort separately showed that M-CLL carrying the IGLV3-21R110 had a shorter TTFT than M-CLL lacking this mutation (P < .02) and similar to U-CLL (P > .12) (supplemental Figure 5). Regarding epigenetic subtypes, patients with i-CLL carrying IGLV3-21R110 had a shorter TTFT compared with i-CLL lacking this mutation (P < .01) and similar to naïve-like CLL (n-CLL; P > .1). Contrarily, i-CLL lacking IGLV3-21R110 had a TTFT similar to that of memory-like CLL (m-CLL; P > .5) (Figure 1E).
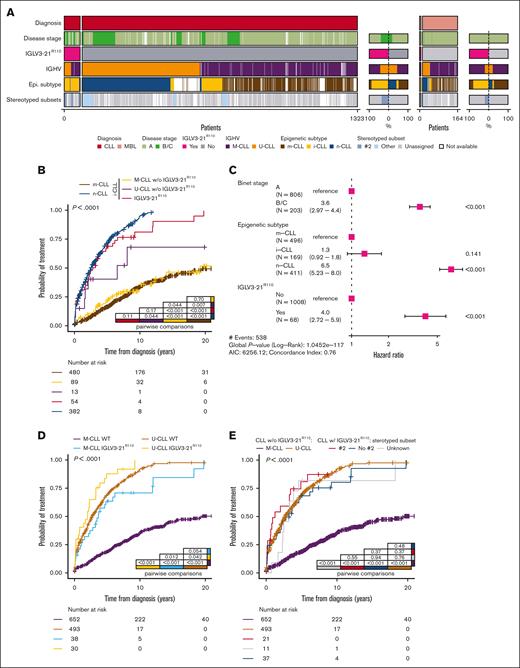
We then combined these novel data with the previously published results of 489 independent patients from our International Cancer Genome Consortium (ICGC)-CLL cohort (54 MBL and 435 CLL)6 and performed analyses on a merged cohort of 1487 patients with IGLV3-21R110 status available (1323 CLL and 164 MBL) (Figure 1B; supplemental Figure 6). The IGLV3-21R110 mutation was present in 78 of 1487 (5.2%) patients and was similarly distributed in CLL (73/1323, 5.5%) and MBL (5/164, 3.05%) (P = .26). IGLV3-21R110 CLL encompassed both M-CLL (41/72, 56.9%) and U-CLL (31/72, 43.1%) (note that IGHV status was not available in 1 patient); whereas, 58 of 68 (85.2%) were classified as i-CLL (epigenetic subtype was not available in 5 IGLV3-21R110 CLL). All stereotyped subset #2 tumors (N = 23; 22 CLL, 1 MBL) carried IGLV3-21R110 but 42 of 66 (65.1%) IGLV3-21R110 CLL were not subset #2 (stereotypy was not available in 12 patients) (Figure 2A). No statistical differences were found in the distribution of genomic alterations in IGLV3-21R110 CLL classified as subset #2 compared with nonstereotyped IGLV3-21R110 cases (supplemental Figure 7).
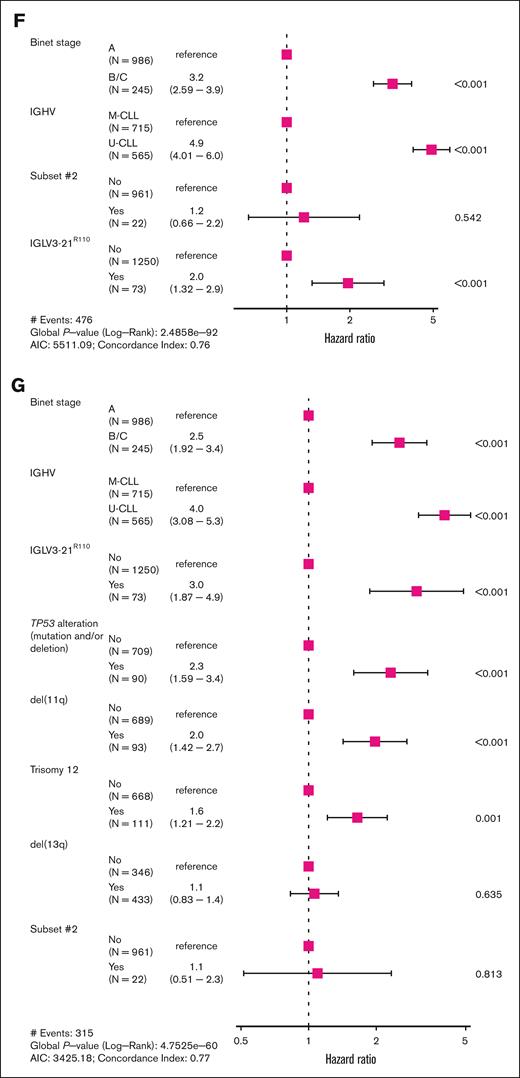
Compared with previous studies in smaller cohorts,5,6 clinical analyses in this integrated cohort provided a higher resolution to study the prognostic value of IGLV3-21R110 within CLL subtypes. Regarding the epigenetic classification of CLL, we confirmed6 that the presence of the IGLV3-21R110 stratifies patients with i-CLL into 2 subgroups with significant differences in their TTFT (P < .001), with i-CLL carrying the IGLV3-21R110 following a disease course similar to n-CLL (P = .11), whereas i-CLL lacked the IGLV3-21R110 similar to m-CLL (P = .23) (supplemental Figure 8). Notably, i-CLL lacking the IGLV3-21R110 and classified as U-CLL had a shorter TTFT compared with i-CLL lacking the IGLV3-21R110 belonging to M-CLL (P = .04) and m-CLL (P = .007). Contrarily, i-CLL lacking IGLV3-21R110 and classified as M-CLL had a TTFT virtually identical to that of m-CLL (P = .7) (Figure 2B). In the multivariable model, IGLV3-21R110 retained prognostic value for TTFT independent of disease stage and n-CLL subtype, whereas the i-CLL subtype did not retain independent prognostic value (Figure 2C). Regarding IGHV subtypes, we found that IGLV3-21R110 had a prognostic impact on M-CLL and U-CLL (Figure 2D). M-CLL carrying the IGLV3-21R110 had a significantly shorter TTFT compared with M-CLL lacking the mutation (P < .001) but a trend toward a longer TTFT that U-CLL (P = .054). In contrast, U-CLL harboring IGLV3-21R110 had a shorter TTFT than U-CLL lacking this mutation (P = .04) (Figure 2D). Considering that 22 of 61 (36%) CLL carrying the IGLV3-21R110 belonged to stereotyped subset #2, we performed an analysis stratifying IGLV3-21R110 CLL based on their stereotypy. All IGLV3-21R110 CLL had a similar TTFT independent of belonging to stereotyped subset #2 (Figure 2E; supplemental Figure 9). In line with these results, IGLV3-21R110 had prognostic value in a multivariate analysis, independent of the disease stage and IGHV gene SHM status, whereas stereotyped subset #2 lost its independent prognostic value (Figure 2F; supplemental Figure 10). IGLV3-21R110 also retained its prognostic value independent of TP53 mutations/deletions, trisomy 12, and deletions in 11q and 13q (Figure 2G).
Overall, we developed a reliable msPCR assay suitable for IGLV3-21R110 screening in a large cohort of patients, allowing us to study the IGLV3-21R110 mutation in 1487 patients. We found new associations between IGLV3-21R110 and other CLL driver alterations while refining the prognostic value of this mutation within CLL subtypes. Our results confirm that IGLV3-21R110 recognizes a subgroup of CLL with aggressive clinical behavior independent of the epigenetic subtypes, IGHV gene SHM status, and stereotyped subset #2, being stereotyped subset #2 prognostically irrelevant for TTFT once considered IGLV3-21R110. Notably, patients with IGLV3-21R110 CLL may benefit from targeted therapies9 that are detrimental to chemoimmunotherapy regimens.5,6 The msPCR assay developed here, as well as similar PCR-based methods recently described,9,16 might help to characterize IGLV3-21R110. Nonetheless, including the IGLV3-21R110 and IGHV gene SHM testing in gene panels may result in a 1-test, integrative next-generation sequencing solution that would indeed facilitate the routine (immuno)genomic characterization of CLL. Overall, this study confirmed that IGLV3-21R110, but not stereotyped subset #2, fully recognized a clinically aggressive subgroup of CLL.
Acknowledgments: The authors thank the technical support of SOPHiA GENETICS, Diagnóstica Longwood, the Genomics Core Facility, the Flow Cytometry and Cell Sorting Core Facility of the Fundació de Recerca Clínic Barcelona-Institut d’Investigacions Biomèdiques August Pi i Sunyer (FCRB-IDIBAPS), and the Hematopathology Collection registered at the Biobank of the Hospital Clínic - FCRB-IDIBAPS. This study was supported by the “la Caixa” Foundation (CLLEvolution - LCF/PR/HR17/52150017 [HR17-00221LCF] and CLLSYSTEMS - LCF/PR/HR22/52420015 [HR22-00172] Health Research 2017 and 2022 Programs; E. Colado), the Ministry of Science and Innovation (MCIN/AEI/10.13039/501100011033) and the European Regional Development Fund “Una manera de hacer Europa” (PID2021-123054OB-I00; E. Campo), European Union (EU) NextGenerationEU/Mecanismo para la Recuperación y la Resiliencia and the Instituto de Salud Carlos III (PMP15/00007; E. Campo), the European Research Council under the EU’s Horizon 2020 Research and Innovation Program (810287, BCLLatlas; E. Campo and J.I.M.-S.), the Generalitat de Catalunya Suport Grups de Recerca AGAUR (2017-SGR-1142 and 2021-SGR-01172; E. Campo), the National Institutes of Health/National Cancer Institute grant P01 CA206978 (C.J.W. and G.G.), and the Centro de Investigación Biomédica en Red de Cáncer. H.P.-A. is a recipient of a predoctoral fellowship from the Spanish Ministry of Universities (FPU19/03110). M.D.-F. acknowledges research support from the Asociación Española Contra el Cáncer Scientific Foundation. B.A.K. was supported by a long-term European Molecular Biology Organization fellowship (ALTF 14-2018). C.K.H. was supported by the National Heart, Lung, and Blood Institute Training Program in Molecular Hematology (T32HL116324). F.N. acknowledges research support from the American Association for Cancer Research (2021 AACR-Amgen Fellowship in Clinical/Translational Cancer Research, 21-40-11-NADE), European Hematology Association (EHA Junior Research grant 2021, RG-202012-00245), and Lady Tata Memorial Trust (International Award for Research in Leukaemia 2021-2022, LADY_TATA_21_3223). E. Campo is an academia researcher of the “Institució Catalana de Recerca I Estudis Avançats” of the Generalitat de Catalunya. This work was partially developed at the Centre Esther Koplowitz (Barcelona, Spain).
Contribution: C.S. contributed to the design of the study, performed the experiments, analyzed and interpreted the data, and wrote the manuscript; B.P.-B. performed the experiments and analyzed and interpreted the data; N.R. contributed to sample preparation and performed the experiments; H.P.-A. performed calcium-flux analysis; T.B., M.D.-F., M.K., A.C.-M., P.M., M.A., M.G., A.N.-B., E. Colado, A.R.P., M.A., M.J.T., J.L., B.A.K., C.K.H., S.R.-G., A.E., C.J.W., G.G., T.Z., A.L.-G., J.I.M.-S., D.C., and J.D. provided samples and data, contributed to experiments, and/or analyzed the data; E. Campo contributed to the study design and manuscript preparation; F.N. designed and supervised the study, performed the experiments, analyzed and interpreted the data, and wrote the manuscript; and all authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: C.J.W., G.G., B.A.K., and C.K.H. are inventors on a patent “Compositions, panels, and methods for characterizing chronic lymphocytic leukemia” (PCT/US21/45144). E. Campo has been a consultant for Takeda, NanoString, AbbVie, and Illumina; has received honoraria from Janssen, EUSA Pharma, and Roche for speaking at educational activities; has received research funding from AstraZeneca; and is an inventor of 2 patents filed by the National Institutes of Health, National Cancer Institute: “Methods for selecting and treating lymphoma types,” licensed to NanoString Technologies, and “Evaluation of mantle cell lymphoma and methods related thereof,” not related to this project. F.N. received honoraria from Janssen, AbbVie, and SOPHiA GENETICS for speaking during educational activities. E. Campo and F.N. licensed the use of protected IgCaller algorithm for Diagnóstica Longwood. The remaining authors declare no competing financial interests.
The current affiliation for T.B. is Hospital Universitario 12 de Octubre, Madrid, Spain.
Correspondence: Ferran Nadeu, Molecular Pathology of Lymphoid Neoplasms, Institut d’Investigacions Biomèdiques August Pi i Sunyer, Rosselló 153, 08036 Barcelona, Spain; e-mail: nadeu@recerca.clinic.cat.
References
Author notes
All data and protocols are provided in the supplemental Material.
Further details are available on request from the corresponding author, Ferran Nadeu (nadeu@recerca.clinic.cat).
The full-text version of this article contains a data supplement.