In this issue of Blood Advances, Syrykh et al1 developed a multiplex polymerase chain reaction (PCR)–based assay to rapidly identify the IGLV3-21R110 mutation and by characterizing independent, treatment-naïve cohorts, highlight its strong negative prognostic impact in chronic lymphocytic leukemia (CLL).
Since more than 2 decades, the somatic hypermutation (SHM) status of the immunoglobulin heavy variable (IGHV) gene divides CLL into 2 clinically relevant subgroups, that is, patients with unmutated IGHV genes (U-CLL) who have a poor prognosis (30%-40% of patients, >98% identity to germline) and patients with mutated IGHV genes (M-CLL) who have a good prognosis (60%-70% of patients, ≤98% identity to germline; see figure).2 In numerous studies throughout the years, the IGHV gene SHM status has remained as one of the strongest prognostic markers in CLL. In more recent years, it has become evident that the IGHV gene SHM status is also a predictive marker because patients with U-CLL show a very high response rate to targeted therapy (Bruton tyrosine kinase and B-cell lymphoma 2 inhibitors) and superior outcome compared with chemoimmunotherapy. For this reason, the IGHV gene SHM status was introduced in the latest International Workshop on CLL guidelines as a mandatory marker to be evaluated before the start of treatment for all patients with CLL.3 To standardize the analysis, the European Research Initiative on CLL has provided recommendations regarding how to assess the IGHV gene SHM status and has also set up an international certification system.4 Notably, this analysis only needs to be performed once because the SHM status does not change over time.
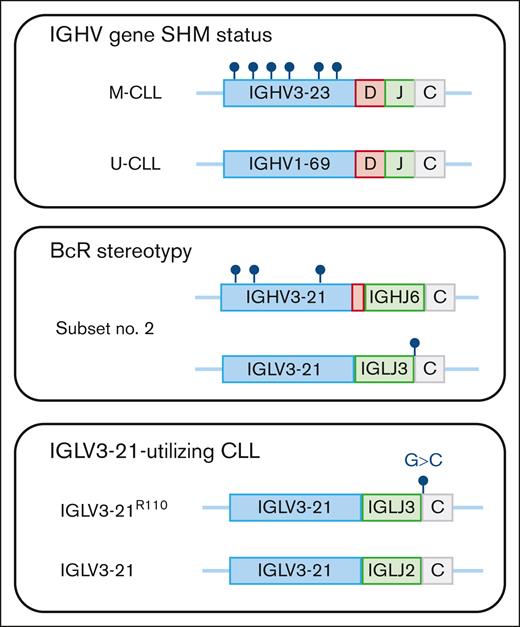
Immunogenetic analysis in CLL. Sequencing of the clonotypic IGHV gene rearrangement identifies whether patients with CLL carry IGHV gene somatic hypermutations (M-CLL) or show few or no somatic hypermutations (U-CLL). By analyzing the IGHV-D-J gene usage and the composition of the CDR3, patients with CLL with stereotyped BcR immunoglobulins can be recognized. Here, exemplified by subset number 2 (IGHV3-21/IGLV3-21), which often shows borderline mutation status and always carries the IGLV3-21R110 mutation. Finally, the IGLV3-21R110 mutation, which occurs at the border between the IGLJ and IGLC gene, can be assessed by performing a separate multiplex PCR reaction as that proposed by Syrykh et al.
Immunogenetic analysis in CLL. Sequencing of the clonotypic IGHV gene rearrangement identifies whether patients with CLL carry IGHV gene somatic hypermutations (M-CLL) or show few or no somatic hypermutations (U-CLL). By analyzing the IGHV-D-J gene usage and the composition of the CDR3, patients with CLL with stereotyped BcR immunoglobulins can be recognized. Here, exemplified by subset number 2 (IGHV3-21/IGLV3-21), which often shows borderline mutation status and always carries the IGLV3-21R110 mutation. Finally, the IGLV3-21R110 mutation, which occurs at the border between the IGLJ and IGLC gene, can be assessed by performing a separate multiplex PCR reaction as that proposed by Syrykh et al.
Already in the 1990s, it became evident that patients with CLL may express B-cell receptor (BcR) immunoglobulins that resemble each other with similar IGHV-IGHD-IGHJ gene usage as well as sequence similarity within the complementarity-determining region 3 (CDR3), the most hypervariable region spanning the IGHV-IGHD-IGHJ junction. One of the early findings was the discovery of patients utilizing the IGHV3-21 gene with IGLV3-21 light-chain usage and a similar, short CDR3 sequence.5 This patient group was later named stereotyped subset number 2 and was shown to display a dismal outcome independent of IGHV gene SHM status (see figure). Today, stereotyped BcR immunoglobulins are detected in >40% of all patients with CLL, and numerous subsets have been identified.6
A unique phenomenon in CLL is that the BcR immunoglobulins can engage in BcR-BcR interaction, triggering cell-autonomous BcR signaling, the so-called homeotypic interaction. A few years ago, several research teams decided to look into the role of homeotypic interaction in subset number 2 and identified a somatic hypermutation, R110, in the junction between the IGHJ and IGHC gene that was critical for this type of interaction (see figure).7 Although the researchers could show that all subset number 2 patients carried this mutation, the R110 mutation was also identified among other patients with IGLV3-21-utilizing CLL without stereotyped BcR immunoglobulins. These patients demonstrated a worse outcome compared with patients with non-R110 IGLV3-21-utilizing CLL. This finding was verified by independent groups, and it became apparent that a new poor-prognosis marker had been discovered.8,9
In the study by Syrykh et al, they developed a PCR-based assay to easily identify the IGLV3-21R110 mutation and demonstrated a 100% concordance using previously characterized CLL cases with a known IGLV3-21R110 mutation status. By analyzing 2 CLL cohorts including 622 (cohort 1) and 376 (cohort 2) patients who were treatment naïve, they identified 3.4% and 7.7% of patients, respectively, in each cohort carrying this mutation. IGLV3-21R110-utilizing CLL demonstrated borderline IGHV identity (97%-97.9% identity), was enriched in the epigenetic intermediate CLL (i-CLL) subtype, and SF3B1 and ATM were more frequently mutated, as previously reported.8,9 As a novel finding, they identified an enrichment of NOTCH1 mutations in IGLV3-21R110-utilizing CLL.
Survival analysis in both cohorts showed that patients with M-CLL harboring the IGLV3-21R110 mutation had a considerably shorter time to first treatment (TTFT) compared with patients with M-CLL without the R110 mutations. As a next step, they pooled the 2 cohorts with their previously characterized International Cancer Genome Consortium (ICGC)–CLL cohort, totaling 1487 patients. Overall, both U-CLL and M-CLL cases carrying the R110 mutation had a worse outcome compared with their wild-type counterparts. Notably, patients belonging to the i-CLL epitype carrying the R110 mutation had a shorter TTFT, similar to naïve-like CLL. In contrast, i-CLL without the R110 mutation demonstrated a TTFT similar to memory-like CLL.
Taken together, this study reinforces the clinical importance of investigating the IGLV3-21R110 mutation status to identify patients at high risk. Although one-third of R110 mutations were observed in subset number 2, the remaining two-thirds did not carry stereotyped BcR immunoglobulins, at least among the 19 stereotyped subsets investigated. Hence, it will not be enough to assess for BcR stereotypy to identify these patients. The proposed PCR assay herein, and by others, could be incorporated in the diagnostic setup of CLL, once standardized, in parallel to the IGHV gene SHM status. Nevertheless, and as raised in this paper, the possibility to incorporate the assessment of both the IGHV gene SHM and the IGLV3-21R110 status would be feasible using next-generation sequencing–based gene panels. In fact, the authors have already published a separate study based on capture-based sequencing in which oncogenic aberrations were assessed in parallel to immunogenetic features.10 Finally, in a few retrospective studies, it has been demonstrated that subset number 2 patients have a dismal outcome if treated with chemoimmunotherapy.11 Therefore, a question that remains is whether patients with a stereotyped R110 mutation would respond better to targeted therapy? To address this question, future large-scale studies will be necessary to reach sufficient numbers to draw solid conclusions. That being said, based on this study and others, it is time to discuss whether the assessment of the IGLV3-21R110 status should be incorporated in the prognostic workup of CLL.
Conflict-of-interest disclosure: R.R. received honoraria from AbbVie, AstraZeneca, Janssen, Illumina, and Roche.