Key Points
PMBL cases with high gene expression of both PDL1 and PDL2 represent a subset of 30% of the population.
These patients have strong immune privilege and poorer outcomes.
Abstract
Primary mediastinal B-cell lymphoma (PMBL) is an uncommon entity of aggressive B-cell lymphoma with an unusually good prognosis, except for 10-15% of chemotherapy-refractory cases. To identify earlier these higher risk patients, we performed molecular characterization of a retrospective multicenter cohort of patients treated with firstline immunochemotherapy. The traits of the patients with gene-expression profiling data (n = 120) were as follows: median age of 34 years (range, 18-67 years); female sex, 58.3%; elevated lactate dehydrogenase, 82.5%; Eastern Cooperative Oncology Group performance status score of 0 to 1, 85.7%; Ann Arbor stage I/II, 55%; International Prognostic Index score of 1 to 2, 64.4%; and median metabolic tumor volume, 290.4 cm3 (range, 15.7-1147.5 cm3). Among all 137 markers tested for correlation with survival data, only programmed death-ligand (PDL) 1 and PDL2 expression showed a prognostic impact. Overall, both PDL1 and PDL2 genes were highly expressed in 37 patients (30.8%; PDL1high/PDL2high). The baseline clinical characteristics of patients with PDL1high/PDL2high were similar to those of other patients. In univariate analysis, PDL1high/PDL2high status was associated with poor progression-free survival (PFS) (hazard ratio [HR], 4.292) and overall survival (OS; HR, 8.24). In multivariate analysis, PDL1high/PDL2high status was an independent prognostic factor of adverse outcomes (PFS: HR, 5.22; OS: HR, 10.368). We validated these results in an independent cohort of 40 patients and confirmed the significant association between PDL1high/PDL2high status and inferior PFS (HR, 6.11). High PDL1/PDL2 gene expression defines a population with strong immune privilege and poorer outcomes from standard chemotherapy who might benefit from firstline checkpoint inhibitor therapy.
Introduction
Primary mediastinal B-cell lymphoma (PMBL) has been recognized since 2001 as a distinct entity classified by the World Health Organization classification as a mature aggressive large B-cell lymphoma (LBCL) of assumed thymic B-cell origin that commonly presents as a large tumor mass in the anterior mediastinum.1,2 The prevalence of PMBL is low, accounting for only ∼5% to 10% of aggressive LBCLs. PMBL predominantly occurs in young female patients (sex ratio, 1.5), but the reason for this particular demographic feature remains unclear. It may be challenging to distinguish PMBL from systemic diffuse large B-cell lymphoma (DLBCL) with secondary mediastinal involvement as well as from mediastinal gray-zone lymphoma.3,4 Routine diagnosis relies on clinical presentation (large anterior mediastinal mass, predominantly in young female patients) and standard histology. The 2022 World Health Organization classification defined essential and desirable criteria for PMBL diagnosis2; essential criteria: (1) presence of an LBCL in the anterior mediastinum and (2) mature B-cell immunophenotype accompanied by at least partial expression of CD23 and CD30; desirable criteria: (1) distinctive stromal sclerosis; (2) expression of at least 1 of the markers MAL, CD200, programmed death-ligand (PDL) 1, and PDL2; and (3) copy gain or rearrangement of the PDL1/PDL2 locus and/or rearrangement involving CIITA. Several recent studies precisely depicted the biology of PMBL, with highly recurrent oncogenic mutations in the Janus kinase–signal transducer and activator of transcription (JAK-STAT) and nuclear factor κB pathways as well as frequent B2M alterations that limit major histocompatibility complex class I expression, but no correlation was observed between these factors and patient outcome.5-7 In general, available cytogenetic and molecular biology tools, such as PDL1/PDL2 or CIITA fluorescence in situ hybridization (FISH),8,9 gene-expression profiling (GEP), and next-generation sequencing (NGS) assays,6,10-12 are not routinely included in the diagnostic procedures for PMBL at the majority of centers.
We recently reported on a cohort of 313 patients with newly diagnosed PMBL who were retrospectively evaluated in a multicenter study by the Lymphoma Study Association (LYSA).13 We observed a strong prognostic impact of baseline total metabolic tumor volume (MTV), with MTV ≥ 360 cm3 being associated with lower progression-free survival (PFS; hazard ratio [HR], 2.18; 95% confidence interval [CI], 1.05-4.53) and overall survival (OS; HR, 4.26; 95% CI, 1.5-12.1), independent of treatment modality and International Prognostic Index (IPI) score. However, the biological substratum for the MTV spectrum and the importance of treatment dose intensity in PMBL remains unknown.14 To date, no risk factor for PMBL-specific lymphomagenesis has been demonstrated.
To better describe our PMBL cohort and to demonstrate the relevance of molecular biology assessment to PMBL diagnosis, with the goal of identifying patients at high risk of chemotherapy resistance earlier, we collected all available diagnostic biopsy data and performed molecular characterization. The aim of this study was to describe the biological features of this cohort and to establish correlations with patient outcomes based on GEP and NGS data.
Methods
Patients and data collection
The PMBL LYSA cohort was a multicenter, retrospective study that assessed the clinical outcomes of patients with PMBL who were previously untreated and received 3 standard immunochemotherapy approaches with or without radiotherapy.13 Briefly, the inclusion criteria were as follows: treatment-naïve adult patients with PMBL diagnosis established at each center, firstline treatment with doxorubicin, cyclophosphamide, vindesine, bleomycin, and prednisone (ACVBP) or cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) plus anti-CD20 between 2007 and 2017, available pretreatment (baseline) positron emission tomography data, and a patient nonopposition statement. In total, 313 patients were enrolled from 25 LYSA centers and received rituximab (R) plus either ACVBP (n = 180; 57.5%) or CHOP delivered every 14 days (R-CHOP14; n = 76; 24.3%) or 21 days (R-CHOP21; n = 57; 18.2%). Available formalin-fixed, paraffin-embedded (FFPE) tissue blocks, fresh-frozen tissue, and immunohistochemistry (IHC) slides obtained at the time of initial diagnosis were collected at the LYSA Pathology Department, Henri Mondor Hospital, Paris, France. The LYSA scientific committee (January 2018), the participating LYSA centers, and the Centre Henri Becquerel Internal Review Board (no. 1916B) approved the study. The study was conducted in accordance with the criteria set by the Declaration of Helsinki.
Expert pathologic review
A histopathologic central review (including several meetings) of all cases with available material was performed by 4 expert hematopathologists (A.T.-G., T.J.M., E.-L.V., and F.D.) following the diagnostic criteria established by previous pathologic descriptions of PMBL from the literature and international classifications1,2,15 (supplemental Methods). Tissue microarrays (TMAs) containing 2 1-mm cores per case were constructed using standard techniques.
GEP
An assay combining reverse transcriptase multiplex ligation-dependent probe amplification and NGS (RT-MLPSeq) was applied as previously described.11 This tool uses a panel of 137 gene expression markers (LymphoSign signature, Genexpath, Rouen, France) to evaluate expression of B-cell differentiation markers, therapeutic targets, prognostic markers, T-cell and macrophage markers, and genes involved in the antitumor immune response (supplemental Methods).11
NGS assay
Somatic variant analysis was performed using a custom 45-gene NGS panel (supplemental Table 1). Briefly, genomic libraries were prepared using an input of 40 ng of fresh-frozen or FFPE DNA and the amplicon-based Qiaseq targeted DNA panel library kit (Qiaseq targeted DNA panel, Qiagen, Hilden, Germany) with unique molecular identifier according to the manufacturer’s instructions. Indexed and target-enriched libraries were pooled and sequenced with an Illumina NextSeq550 using 150–base pair paired-end reads (Illumina, San Diego, CA). Raw sequencing signals were analyzed using the Illumina platform, and were aligned to the hg19 reference genome. Variant calling was performed by unique molecular identifier Varcal software (supplemental Methods; supplemental Figure 1).16
FISH
FISH analysis for detection of 9p24.1 gains, amplifications, and chromosomal translocations was performed using standard methods with TMA slides (interphase FISH) and metaphase chromosomes from available cultured tumoral biopsy samples (supplemental Methods; supplemental Figure 2).
Study end points
The 2 primary end points were PFS and OS for the patients who underwent GEP analysis. Secondary end points were as follows: (1) a description of the morphological, immunophenotypical, and molecular spectrum; (2) a description of the GEP and NGS profiles according to baseline MTV (cutoff ≥ 360 cm3); and (3) PFS and OS for whom NGS data were available.
Validation cohort
To validate our analyses of PFS and OS in the GEP set, we performed the RT-MLPSeq assay on an independent monocentric cohort of patients routinely treated with frontline R-ACVBP (n = 20), R-CHOP14 (n = 11), or R-CHOP21 (n = 9) for PMBL at the Centre Henri Becquerel between 2007 and 2022, with available RNA samples extracted from diagnostic tumor biopsy samples (n = 40). PMBL diagnoses were locally established based on international classifications1,2,15 by experienced hemopathologists from the Lymphopath network.17 These patients were not included in the PMBL LYSA cohort either because positron emission tomography examination before treatment was not available or because they were diagnosed after the inclusion period for the PMBL LYSA cohort.
Statistical analysis
Patient and tumor characteristics are described with median and extreme values for quantitative variables and numbers and percentages for qualitative variables. Statistical differences in some parameters between different subgroups of patients were determined using the χ2 teste (or Fisher exact test when appropriate) for qualitative variables, and the Wilcoxon Mann Whitney U test for nonnormal quantitative variables. To take into account multiple comparisons, we carried out false discovery rate correction, and we present the corresponding adjusted P values in tables. OS was calculated as the number of months from the biopsy date (that determined PMBL diagnosis) to the date of death or last follow-up date. PFS was calculated in months from the biopsy date to the date of progression, death, or last follow-up (supplemental Methods). P values and adjusted P values < .05 were considered statistically significant. All statistical analyses were performed with R software version 4.0.0.
Results
Patient characteristics and expert pathologic review
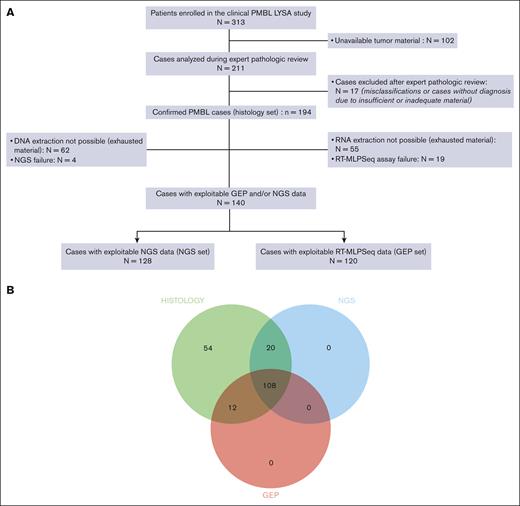
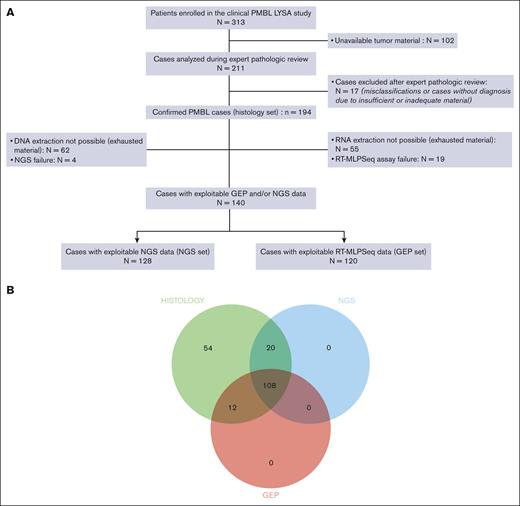
From the baseline PMBL LYSA cohort of 313 patients, tumor material was received at the LYSA Pathology Department for 211 cases (67.4%) from 22 centers (supplemental Table 2). During the expert pathologic review, 17 of 211 cases (8%) were excluded from all further analyses because of misclassifications or a lack of diagnosis because of insufficient or inadequate material (supplemental Table 3). Overall, 194 confirmed PMBL cases (of 313 patients; 62% of the initial cohort) were included in this study (the so-called histology set). Patients in the histology set had the following traits: median age of 33 years (range, 18-68 years); female sex, 59.8%; elevated lactate dehydrogenase level, 83%; Eastern Cooperative Oncology Group performance status score of 0 to 1, 84.7%; Ann Arbor stage I to II, 56.7%; IPI score of 1 to 2, 63.3%; median MTV, 300.4 cm3 (range, 2.5-1403.6 cm3); and mediastinal involvement: 96.8% (Table 1). Details of the morphology and IHC results are summarized in supplemental Table 4. Not all IHC experiments with all antigens could be performed for all cases because of progressive exhaustion of the tumor material available. Typically, PMBL displayed the following immunophenotype: CD20+ (99.5%), CD15− (98.8%), CD30+ (88.5%; weak and heterogeneous staining: 87.2%), CD23+ (73.4%), BCL2+ (92.6%), CD10− (84.8%), BCL6+ (97.2%), MUM1+ (90.5%), PDL1+ (78%), and PDL2+ (50%). supplemental Table 5 summarizes the nucleic acid concentrations and the RT-MLPSeq quality score. NGS and GEP failed for 4 (3%) and 19 (13.7%) samples, respectively. Patients with GEP data (n = 120) were included in the GEP set (62% of the histology set), and patients with NGS data (n = 128) were included in the NGS set (66% of the histology set; Figure 1A). Overall, data from at least 1 of the 2 assays were obtained for 140 patients (72%), with both types of data available for 108 patients (56%) (Figure 1B). No differences in patient characteristics were observed between the 2 patient sets (supplemental Table 6).
Study overview. (A) Study flowchart. (B) Venn diagram representing the different subcohorts of the study (histology set: n = 194; NGS set: n = 128; and GEP set: n = 120).
Study overview. (A) Study flowchart. (B) Venn diagram representing the different subcohorts of the study (histology set: n = 194; NGS set: n = 128; and GEP set: n = 120).
NGS mutational landscape
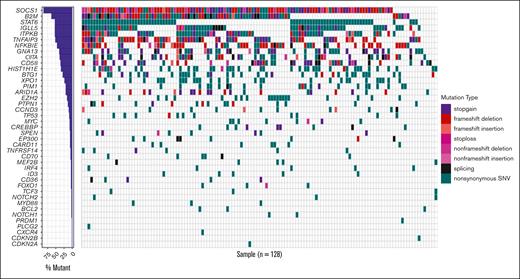
Among the 128 patients with available tumor DNA sequencing data, the median (q1 and q3) depth of coverage was 2992 times (2490 and 3453, respectively); all patients carried at least 1 detectable variant, that is, there was an informative rate of 100% of the 45-gene panel (supplemental Table 7). The genes mutated in >20% of the patients (Figure 2) were as follows: SOCS1 (87.5%), B2M (63.3%), STAT6 (53.1%), IGLL5 (53.1%), ITPKB (52.3%), TNFAIP3 (51.6%), NFKBIE (46.1%), GNA13 (39.8%), CIITA (38.3%), CD58 (35.2%), HIST1H1E (34.4%), BTG1 (28.1%), and XPO1 (22.7%). We detected a median of 15 (range, 1-52) mutations per case, without mutations in BRAF, BTK, CD79A, or CD79B. Of note, we observed 3 cases with MYD88 nonsynonymous mutations that were not the classical L265P hot spot variant (p.218S: n = 1, p.S161C: n = 1, and p.S164C: n = 1; supplemental Table 7) and 20 cases with EZH2 nonsynonymous mutations, including 18 of the classical Y646 hot spot variant (supplemental Table 8).
Heat map representing the mutational landscape of the PMBL cohort. SNV, single nucleotide variant.
Heat map representing the mutational landscape of the PMBL cohort. SNV, single nucleotide variant.
Correlation of molecular biology with metabolic imaging
We obtained baseline MTV data for 148 patients (76.3%) from the histology set. To identify the biological substratum for the MTV spectrum, we evaluated variation in PMBL molecular profiles (NGS and GEP) between patients with high (≥360 cm3) and low (<360 cm3) MTVs. We observed that NGS data for patients with a high baseline MTV (n = 33) displayed a different somatic mutational pattern compared with NGS data for patients with low baseline MTVs (n = 62), with a twofold to fivefold higher rate of specific somatic alterations targeting several genes, including TP53, FOXO1, EZH2, NOTCH1, and CXCR4 (supplemental Table 9). In contrast, the GEP data for patients with MTV ≥360 cm3 (n = 40) were not significantly different from those for GEP patients with low MTV (supplemental Table 10).
Correlation of GEP data with patient outcomes
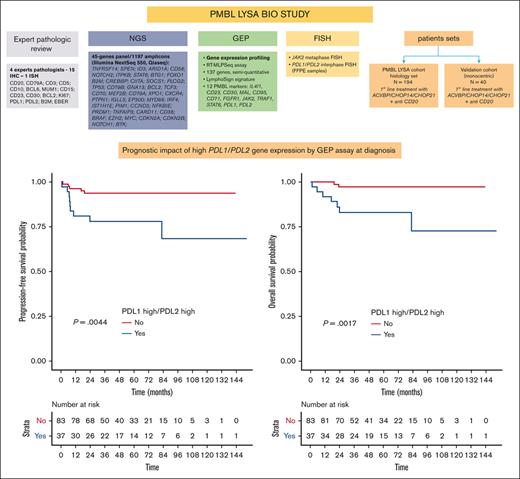
Among all 137 markers tested for correlation with survival data, only PDL1 and PDL2 expression levels had a prognostic impact. Indeed, patients with high PDL1 gene expression (ie, cases with high PDL1 expression; cutoff: median expression of PDL1 = 0.402) had inferior outcomes (PFS: HR, 6.66; 95% CI, 1.49-29.8 [P = .0041]; OS: HR, 8.63; 95% CI, 1.08-69.1 [P = .014]), as did patients with high PDL2 gene expression (cutoff: median expression of PDL2 = 9.147; PFS: HR, 2.52; 95% CI, 0.79-8.04; P = .11; OS: HR, 8.12; 95% CI, 1.02-64.9; P = .018; supplemental Figures 3 and 4). Within the GEP set (n = 120), 37 patients (30.8%) showed high expression of both PDL1 and PDL2 genes (so-called PDL1high/PDL2high cases; cutoff: median expression of each gene).
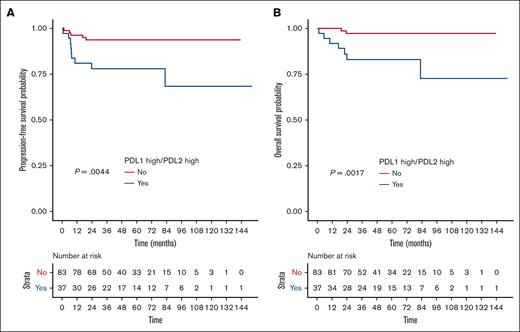
Overall, the 2-year PFS rate in the PDL1high/PDL2high group was estimated at 78.1% vs 93.8% for other patients (HR, 4.292; 95% CI, 1.447-12.817; P = .0044), and the 2-year OS rate in the PDL1high/PDL2high group was estimated at 83.2% vs 97.3% for other patients (HR, 8.24; 95% CI, 1.71-39.7; P = .0017; Figure 3). Patients displaying a PDL1high/PDL2low or PDLlow/PDL2high profile had prognoses similar to those of patients with a PDL1low/PDL2low profile (supplemental Figure 5), and PD1 expression had no impact on outcome (data not shown).
High gene expression of both PDL1 and PDL2 correlated with poorer outcome. (A) PFS and (B) OS according to PDL1/PDL2 gene expression status (assessed by RT-MLPSeq) in the GEP set from the PMBL LYSA cohort (n = 120). Within the GEP set (n = 120), we identified a subset of 37 of 120 patients (30.8%) with high gene expression of the PDL1 and PDL2 genes (PDL1high/PDL2high; cutoff: median expression of each gene: PDL1 = 0.402; PDL2 = 9.147).
High gene expression of both PDL1 and PDL2 correlated with poorer outcome. (A) PFS and (B) OS according to PDL1/PDL2 gene expression status (assessed by RT-MLPSeq) in the GEP set from the PMBL LYSA cohort (n = 120). Within the GEP set (n = 120), we identified a subset of 37 of 120 patients (30.8%) with high gene expression of the PDL1 and PDL2 genes (PDL1high/PDL2high; cutoff: median expression of each gene: PDL1 = 0.402; PDL2 = 9.147).
Features of the PDL1high/PDL2high subset
Compared with others, patients with PDL1high/PDL2high had similar baseline characteristics, except for a trend toward a higher proportion of elevated lactate dehydrogenase level (94.6% vs 77.1%; P = .062) and a trend toward higher baseline MTVs (358.5 vs 255.7 cm3; P = .081). Treatments in the PDL1high/PDL2high group did not differ from those in the other groups (Table 2). Mutations in NOTCH2, PRDM1, CDKN2A, IRF4, SPEN, MYD88, NOTCH1, CARD11, and CD58 were enriched (1.5-fold to fivefold higher rate of alterations) in patients with PDL1high/PDL2high (supplemental Table 11), and these patients also displayed a different GEP, with high gene expression of JAK2 and lower expression of CD138 and GATA3 (supplemental Figure 6). By using dual-color metaphase FISH probes targeting JAK2 at 9p24.1 in 17 cases, we observed that patients with PDL1high/PDL2high more frequently exhibited JAK2 copy gains and amplifications (33.3% and 66.7%, respectively) than patients with PDL1low/PDL2low (25% and 0%, respectively) and that patients with PDL1high and/or PDL2high consistently exhibited JAK2 copy gains or amplifications (100% [vs 25% for patients with PDL1low/PDL2low]; weighted κ coefficient of agreement = 0.821 [95% CI, 0.486-1]; supplemental Table 12; supplemental Figure 7). By using 2 different interphase FISH probes targeting PDL1/PDL2 at 9p24.1 in 44 and 47 cases with available FFPE TMAs, we found that patients with PDL1high/PDL2high more frequently showed PDL1/PDL2 copy gains and amplifications (50%-70% and 20%-40%, respectively) than those with PDL1low/PDL2low (31.6%-35.3% and 0%-5.9%, respectively; supplemental Table 13; supplemental Figures 8 and 9). For 35 cases displaying identical results with both interphase FISH probes, patients with PDL1high and/or PDL2high more frequently exhibited PDL1/PDL2 copy gains or amplifications (76% [vs 30% for patients with PDL1low/PDL2low]; weighted κ coefficient of agreement = 0.422 [95% CI, 0.11-0.734]; supplemental Table 13C).
PDL1 and PDL2 protein expression was determined using the tumor proportion score (TPS) (supplemental Figures 10 and 11). Overall, PDL1 and PDL2 TPS results were available for 109 of 194 (56.2%) and 88 of 194 (45.4%) cases, respectively, with GEP results for PDL1 and PDL2 expression available for 90 and 82 cases, respectively. There was a moderate correlation between PDL1 expression and PDL1 TPS (r = 0.43, P < .001; supplemental Figures 12A and 13A) and a good correlation between PDL2 expression and PDL2 TPS (r = 0.69, P < .001; supplemental Figures 12B and 13B) in IHC samples. PDL1 and PDL2 TPS showed no correlation (r = 0.065, P = .781; supplemental Figure 13C). We observed a trend toward inferior PFS for patients with a positive PDL1 TPS (HR, 3.85; [1.11-13.4]; P = .1611), and none of the patients with a negative PDL1 TPS died (supplemental Figures 14 and 15). PDL2 TPS was not associated with outcome (supplemental Figures 16 and 17).
Correlation of NGS data with patient outcomes and features of the B2M-mutated subset
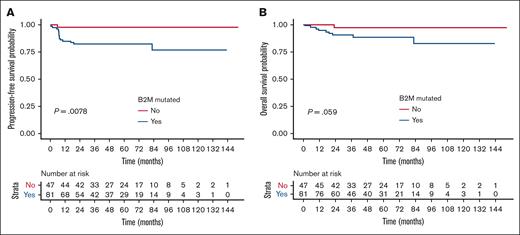
Among all 41 mutated genes tested for correlation with survival data, only B2M mutation had a prognostic impact. Indeed, in univariate analysis, the presence of B2M mutation was associated with lower PFS (HR, 9.455; 95% CI, 1.249-71.595; P = .0078) and a trend toward lower OS (HR, 5.786; 95% CI, 0.732-45.709; P = .059; Figure 4). The B2M mutations (n = 81) detected were mainly inactivating alterations (splicing, stop gains, and frameshift deletions; Figure 2), but mutation status did not correlate with a different level of B2M expression. Compared with others, patients harboring B2M mutation had similar baseline clinical characteristics and received comparable treatments (Table 2) but overexpressed ASB13 (germinal center B-cell like marker) and underexpressed CD56 (supplemental Figure 18). CD56 encodes a protein expressed by many immune cell subsets, including natural killer cells, CD8 T cells, dendritic cells, and monocytes. Overall, 38% of the patients harboring B2M mutation carried several B2M variants. We did not obtain any copy number variation (CNV) data, but the average variant allelic frequency of B2M was higher than the average variant allelic frequency of other somatic mutations in 36% of the patients; this result strongly suggests that B2M mutations are biallelic. B2M mutations had a nonsignificant impact on the proportion of B2M-positive tumor cells by IHC (mean, 26% vs 33% for B2M wild-type cases; P = .424; supplemental Figure 19), and patients with PDL1high/PDL2high displayed a trend toward a lower proportion of B2M-positive tumor cells by IHC (mean, 15% vs 31%; P = .11; supplemental Figures 20 and 21).
B2M mutations correlated with poorer outcome. (A) PFS and (B) OS according to B2M mutation status in the NGS set from the PMBL LYSA cohort (n = 128).
B2M mutations correlated with poorer outcome. (A) PFS and (B) OS according to B2M mutation status in the NGS set from the PMBL LYSA cohort (n = 128).
Univariate and multivariate survival analysis
In univariate analysis, PDL1high/PDL2high status (HR, 4.292; 95% CI, 1.447-12.817; P = .0044), B2M mutation (HR, 9.455; 95% CI, 1.249-71.595; P = .03), and MTV ≥ 360 cm3 (HR, 2.449; 95% CI, 1.06-5.661) were associated with reduced PFS (Table 3). When adjusting PDL1high/PDL2high status effect on PFS with the type of treatment received, the HR remained stable (HR, 4.285; 95% CI, 1.43-12.842; data not shown). We could not perform multivariate analysis for PFS, with 3 of the variables significantly associated with outcome in univariate analysis (MTV, B2M mutation status, and PDL1/PDL2 status) because of a convergence issue of the Cox regression model (related to missing data and few PFS events), leading to nonestimation of the coefficient for B2M. Indeed, the only patient who had a PFS event in the wild-type B2M group was a patient (AB021) for whom it was not possible to measure a baseline MTV. As a consequence, when we adjusted for the 3 variables in the same regression model, the subgroup of patients with wild-type B2M was not part of the subgroup of patients with PFS events. Therefore, multivariate analysis for PFS was performed excluding the B2M variable. The final multivariate Cox regression model thus included PDL1/2 status and MTV, showing PDL1high/PDL2high status to be an independent prognostic factor of inferior PFS (HR, 5.222; 95% CI, 1.352-20.168; P = .03; Table 3).
In univariate analysis, PDL1high/PDL2high status (HR, 8.24; 95% CI, 1.71-39.7; P = .0017), IPI ≥3 (HR, 2.863; 95% CI, 1.135-7.222), and MTV ≥360 cm3 (HR, 4.391; 95% CI, 1.397-13.802; P = .01) were associated with reduced OS. When adjusting PDL1high/PDL2high status effect on OS with the type of treatment received, the HR did not significantly vary (HR, 8.66; 95% CI, 1.769-42.38; data not shown).
Multivariate analysis for OS, including IPI, PDL1/2 status, and MTV, showed that PDL1high/PDL2high status was an independent prognostic factor for reduced OS (HR, 10.368; 95% CI, 1.204-89.267; P = .03; Table 3).
Validation cohort
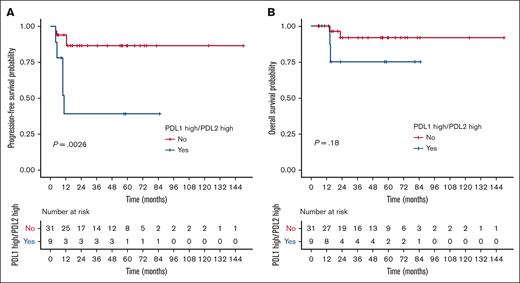
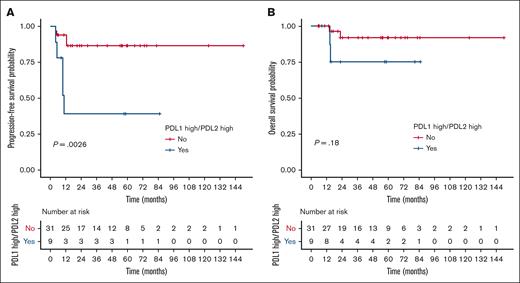
Within the independent cohort of PMBL cases (n = 40; clinical characteristics summarized in supplemental Table 14), we identified a subset of 9 patients (22.5%) with high gene expression of both the PDL1 and PDL2 genes using the same cutoffs as used for the PMBL LYSA cohort. Despite the small number of events (n = 9), we again observed a significant association between PDL1high/PDL2high status and inferior PFS in the validation cohort (HR, 6.11; 95% CI, 1.61-23.2; P = .0026; Figure 5A). Conversely, OS was not significantly affected by PDL1high/PDL2high status (Figure 5B).
Validation cohort for PDL1/PDL2 high gene expression prognostic impact. (A) PFS and (B) OS according to PDL1/PDL2 gene expression status (assessed by RT-MLPSeq) in the independent Centre Henri Becquerel validation cohort (n = 40).
Validation cohort for PDL1/PDL2 high gene expression prognostic impact. (A) PFS and (B) OS according to PDL1/PDL2 gene expression status (assessed by RT-MLPSeq) in the independent Centre Henri Becquerel validation cohort (n = 40).
Discussion
By correlating the GEP of PMBLs with outcomes, we highlight a subset of cases highly expressing both the PDL1 and PDL2 immunosuppressive genes, representing ∼23% to 30% of cases. In this disease, with an overall very good prognosis, we demonstrate, to our knowledge, for the first time, that this group of patients is at high risk of chemotherapy failure because of the specific immune privilege conferred by the high PDL1/PDL2 gene-expression profile. In multivariate analyses, only PDL1high/PDL2high status was an independent prognostic factor of outcomes. Our findings were confirmed in an independent validation cohort, with markedly inferior PFS for patients with PDL1high/PDL2high, despite the small size of the cohort, which emphasizes the robustness of our results. Because almost all of the PMBL cases showed PDL1 and PDL2 gene expression, we used the median expression of each of the genes to isolate the subset expressing these genes at the highest levels while maximizing statistical power, given the low number of progression events in this disease. This cutoff at the median was confirmed in the validation cohort.
Our findings based on GEP experiments were confirmed by FISH analysis, which showed that cases with high gene expression of PDL1/PDL2 mainly arose from genetic gains and amplifications of the 9p24.1 locus. However, the gains and amplifications of this locus are not the only mechanisms explaining the high expression of PDL1/PDL2, because we observed PDL1high/PDL2high cases without genic amplification and cases with 9p24.1 chromosomal translocations without high PDL1/PDL2 gene expression. This suggests the existence of other mechanisms regulating expression of these 2 genes, such as activation of transcription by transcription factors such as MYC and STAT3,18,19 epigenetic modifications under the influence of microRNAs,20 or translational regulation by mechanisms of ubiquitination, deubiquitination, glycosylation, and phosphorylation under the influence of CDK4/621 or CMTM6.22 Overall, our results are globally consistent with prior those of studies from other groups that established the existence of rearrangements, gains, and amplifications of the 9p24.1 locus in DLBCL, nodular-sclerosis classical Hodgkin lymphoma (cHL), and PMBL, with high gene expression of the PDL1/PDL2/JAK2 genes and a correlation with protein expression in IHC experiments. First, Rosenwald et al used GEP and quantitative polymerase chain reaction and observed elevated expression of JAK2, PDL1, and PDL2 at the 9p24.1 locus in more than half of PMBL and cHL cases.23 Then, several teams detected PDL1 expression using flow cytometry and IHC of tumor cells and tumor-infiltrating macrophages (TAMs) in subsets of DLBCL, cHL, and PMBL.24,25 Green et al showed that 38% of nodular-sclerosis cHL and 63% of PMBL cases displayed amplifications of the 9p24.1 locus affecting the PDL1 and PDL2 genes (chr9:5450559-5468477 and chr9:5510545-5571282, respectively, separated by only ∼40 kilobases).26 The amplified 9p24.1 segment includes 977 genes within a 22-megabase region (chr9:1-21944952) and 7 genes within the 177-kilobase amplification peak, including JAK2 (chr9:4985245-5128183). Roosbroeck et al also confirmed that the structural 9p24.1 aberrations in PMBL are hallmarked by a common ∼200-kilobase breakpoint region located downstream of JAK2, harboring PDL1/2, which leads to aberrant expression of PD-1 ligands to promote immune evasion.27 Shi et al used a polymerase chain reaction–based TaqMan copy number assay to detect the copy gain of PDL2 and found that 75% of patients with PMBL but none of the patients with DLBCL had PDL2 gain.28 The authors also observed a strong correlation between PDL2 gains/amplifications and PDL2 staining. Twa et al used FISH for PDL1/PDL2 alterations and observed break-apart and amplification frequencies of 20% and 29% in 125 PMBL cases, respectively, with elevated PDL2 expression by flow cytometry in 3 PDL1/PDL2 rearranged PMBL cell lines. Georgiou et al also used a FISH assay to detect 12% gain and 3% amplification of the PDL1/PDL2 locus in 190 DLBCL samples.8 RNA sequencing data coupled with IHC have revealed that these cytogenetic alterations correlate with increased expression of PDL1 but not of PDL2.29 In 2019, Wang et al also reported the results of a study of alterations of the 9p24.1 locus in DLBCL with a combination of FISH, RNA sequencing, and whole-exome sequencing data. This team showed that ∼10% of DLBCL cases have 9p24.1 rearrangements (gains, amplifications, and translocations), with high expression of PDL1/PDL2 and JAK2, which correlated with IHC staining. These patients were younger, with a female predominance, and had no typical clinical presentation or histology of PMBL but with a longer PFS for cases with 9p24.1 amplification, suggesting that they may be patients with nonmediastinal PMBL-like conditions. In addition, Cheng et al observed that patients with DLBCL with high PDL1 gene expression by GEP (25% of the cases) had shorter OS, and Kiyasu et al reported that patients with DLBCL with PDL1+ tumor cells (11% of the cases) had inferior OS, with no difference in the outcome for microenvironmental PDL1+ staining.30,31
We hypothesize that the PDL1high/PDL2high status is predictive of a response to anti-PD1 immunotherapy, in addition to other mechanisms of sensitivity to PD1 blockade previously described in PMBL by Chapuy et al, such as high tumor mutational burden, microsatellite instability, and apolipoprotéins B mRNA editing enzyme, catalytic polypeptide-like mutational signature.7 Indeed, binding of PDL1 or PDL2 expressed by tumor cells to its receptor PD1 on infiltrating T lymphocytes induces inhibitory signals, thus reducing T-cell cytotoxic activity. This negative modulation provides an immune escape mechanism for tumor cells.32 This PDL1high/PDL2high subset highlights the powerful phenotype of immune privilege; compared with others, these patients with PDL1high/PDL2high probably have a different biology with a more aggressive genotype, including frequent somatic alterations in NOTCH1/2, PRDM1, CDKN2A, IRF4, and CD58, and a different GEP with high gene expression of JAK2. These patients with PDL1high/PDL2high showed no difference of B2M gene expression but displayed a lower expression of B2M protein, further participating in the immune privilege phenotype. The absence of B2M protein hinders the formation of peptide–major histocompatibility complex class I complexes and T-cell stimulation, leading to immune escape and resistance to chemotherapy in lymphomas.33-36 The precise mechanism of B2M expression decrease in PDL1high/PDL2high cases remains to be explored.
Activation of the JAK-STAT pathway is a possible cause of high PDL1/PDL2 gene expression,37 as are PDL1/PDL2 rearrangements and 9p gains. Previous data from studies of DLBCL regarding the impact of PDL1/PDL2 expression have been contradictory. Several teams have demonstrated the prognostic impact of high PDL1 gene expression based on IHC analysis in DLBCL, and others have reported better outcomes for patients with high PDL2 gene expression by IHC38,39 but worse outcomes for those with PDL2 locus amplification40 or elevated soluble PDL1 protein blood levels.41 Rosenwald et al demonstrated that almost all PMBLs expressed high levels of PDL1 and PDL2, with PDL2 being the overall best PMBL distinction gene, with a 5.6-fold higher expression in PMBLs than in DLBCLs.23 Previous studies have found that between 71% and 100% of PMBL tumor cells are PDL1+,24,25 including refractory cases, and that 72% are PDL2+,28 but no correlations with outcome were reported. These findings have paved the way for anti-PD1 development for relapsed/refractory PMBL.42,43 Of note, in the KEYNOTE-013 trial, the objective response rate increased from 25% to 64% in patients with high PDL1 expression by IHC, compared with in others, and a clear association was observed between PDL1/2 copy gains and PDL1 expression. Ansell et al analyzed 9p24.1 alterations in a small fraction of patients with relapsed/refractory DLBCL enrolled in the CheckMate 139 study and receiving nivolumab and reported 3% amplification and 16% gain. Of note, 1 of the 3 patients who achieved complete response had high-level 9p24.1 amplification, whereas the other 2 had normal 9p24.1 copy number or an unavailable biopsy specimen; there was no detectable PDL1 expression on tumor cells from 5 evaluable patients with partial response.44 In our study, we observed good correlation between high PDL1/PDL2 gene expression in GEP and PDL1/PDL2 labeling in IHC, although it was not associated with a difference in outcome based on TPS assessment. As there is no validated cutoff for high PDL1/PDL2 expression by IHC in PMBL, IHC cannot yet serve as a predictive assay of response in these patients.43 The PDL1 and PDL2 expression based on IHC that we observed was almost exclusively related to the tumor cells and not to the microenvironment (MET). We cannot exclude the possibility that the high gene expression of PDL1/PDL2 that we detected in the patients with PDL1high/PDL2high in our series is partly related to expression of PDL1/PDL2 by TAMs. However, we did not observe significant PDL2 staining on MET cells, and Shi et al already demonstrated that 70% of PMBL tumor cells express PDL2, without any labeling of MET cells.28 In addition, the prognostic impact of PDL1 expression by TAMs remains under debate, with some authors suggesting that it is associated with a favorable impact on survival for PMBLs,45 and other authors finding an unfavorable impact on survival for lung cancer.46
The increasing availability of NGS and GEP tools in academic centers suggests the possibility of their routine use in the future. In most academic laboratories, FISH is not routinely used for diagnosis of lymphomas because it is time consuming and requires specific expertise. Indeed, in the study by Laurent et al in the Lymphopath network, it appears that FISH was only used in 9% to 15% of the cases.17 The RT-MLPSeq assay that we used is already commercialized and easily implementable in routine diagnostic laboratories (operating with classic NGS Illumina sequencers with a handling time of approximately half a day). This GEP tool requires low amounts of RNA (50-100 ng in 2 μL) and performs well with FFPE core-needle biopsies. Indeed, the genetic sequences targeted by the probes are particularly short (between 40 and 60 bases), which guarantees very good robustness with respect to RNA degradation; this method is, therefore, particularly suitable for analysis of difficult biological samples such as mediastinal FFPE biopsy specimen. In addition, the finding that not all PDL1high/PDL2high cases correlated systematically with 9p24 amplification by FISH analysis reinforces interest in the companion GEP test because simply performing FISH alone would risk misidentifying the population at higher risk of chemotherapy resistance. Our 45-gene NGS panel showed informativity of 100% (ie, able to detect at least 1 somatic variant in each case of PMBL) and performed well with FFPE DNA samples collected 5 to 15 years prior. The mutational spectrum of PMBL depicted herein, with SOCS1, B2M, and STAT6 being the 3 most frequently mutated genes, is consistent with previous reports in the literature.6,7 We observed that patients with high baseline MTV (≥360 cm3) had a higher rate of somatic alterations affecting FOXO1, TP53, NOTCH1, and BCL2 mutations that were demonstrated to be associated with poor outcome in DLBCL.47-50B2M-mutated cases had worse PFS, consistent with other reports in DLBCL and mediastinal gray-zone lymphoma33-36,51,52 but discordant with another work in cHL.53 Compared with others, B2M-mutated cases displayed reduced expression of CD56, which encodes a protein with a strong role in immune surveillance.54 The role of B2M mutations in predicting response to anti-PD1 treatment is unknown in lymphomas and controversial in solid tumors, with some authors reporting a risk of resistance to anti-PD1 treatment in B2M-mutated lung cancer55 and melanoma,56 and others reporting a benefit from anti–PD-1 therapy in B2M-mutated colorectal carcinoma.57 Application of our NGS panel to the quantification of circulating tumor DNA also seems attractive, given this high level of informativity, with each variant being able to serve as a tracer of the disease for dynamic follow-up of minimal residual disease. A prospective study to explore this topic is ongoing (NCT04824950).
We acknowledge several limitations of this work. First, the lack of availability of diagnostic tumor blocks or their exhaustion (because of insufficient quantity/quality material, mediastinum difficult to biopsy, and frequent needle biopsies), failure of DNA or RNA extraction, and/or insufficient quantity/quality of nucleic acids extracted for molecular biology techniques might have created sampling bias, with many drop-outs from the original PMBL LYSA cohort and cases with complete molecular data comprising a subset of the overall cohort. However, we did not find clinical differences between the different patient subsets and the overall population. Second, the cases were collected over a very long period of time (ie, 10 years, ending in 2017), and 3 different immunochemotherapy regimens were used. However, the distribution of the types of treatment received by patients was similar between the 2 cohorts, with overall ∼55% of patients treated with R-ACVBP, ∼25% with R-CHOP14, and ∼20% with R-CHOP21, and the prognostic effect of PDL1high/PDL2high status was also comparable between the 2 cohorts, limiting the risk of a prognostic confounding effect of the treatment type. We were also able to ensure that the heterogeneity of the treatments did not affect the prognostic effect of the PDL1/PDL2 status by adjusting for this variable in multivariate models containing PDL1/PDL2 status and treatment (data not shown). This is also a reflection of the real-life PMBL setting; indeed, there is no international consensus on the firstline treatment recommended for PMBL, except for use of immunochemotherapy with anthracycline and anti-CD20,58 with recent data confirming that dose-dense regimens without radiotherapy lead to better outcomes.14
Third, we did not have CNV data for our 45-gene NGS panel; thus, we could not correlate the presence of B2M alterations with CNVs of this gene. However, we performed FISH analyses to correlate high PDL1/PDL2 gene expression with amplifications and copy number gains of the 9p24 locus. Our findings confirmed that the PDL1high/PDL2high status by GEP is mainly associated with 9p24.1 amplifications and copy number gains, as previously reported by others in the literature.4,26 However, we could not identify the partner genes involved in the translocations affecting the 9p24.1 locus (cases with JAK2 breaks using metaphase FISH). Finally, we did not perform a mechanistic or functional study to confirm the biological impact of the molecular alterations identified. Regarding our cases with 9p gains and/or amplifications by FISH but with a PDL1high/PDL2low or PDL1low/PDL2high profile by GEP, we hypothesize that this finding might be linked to different characteristics of the 5′ regulatory regions of the 2 genes, particularly at the level of the number of binding sites for transcription factors of the JAK/STAT pathway. Green et al already observed that in the event of amplification of the 9p24.1 locus, the number of PDL2 transcripts was more strongly augmented than that of PDL1 in PMBL.26 Several fundamental studies have reported differences between the mechanisms of the expression and regulation and potentially the functions of the 2 PD1 ligands.59-61 PDL1 and PDL2 do not have the same structure (notably with a 14–amino acid difference in the immunoglobulin variable domain); PDL2 has stronger affinity for PD1, whereas PDL1 has the ability to bind both PD1 and CD80, unlike PDL2, which can only bind PD1.62-64
Anti-PD1 agents are not approved in the firstline setting, probably because PMBLs have an overall very good prognosis and because of the lack of companion tests with predictive value to identify patients who may benefit from anti-PD1 agents. Given our finding of the chemotherapy resistance profile of patients with PDL1high/PDL2high, this suggests that anti-PD1 treatments from diagnosis onward in this subgroup, either as monotherapy or in combination with chemotherapy, should be offered. This population can be easily distinguished by RT-MLPSeq as a diagnostic companion. It has previously been established that immunosurveillance is essential for the eradication of cancers, including lymphomas.65-68 We hypothesize that immunosurveillance mechanisms are markedly impaired in patients with PDL1high/PDL2high PMBL, because of, at least in part, the high expression of these 2 genes; thus, the use of anti-PD1 in combination with chemotherapy might make obtaining a cure possible by stopping the deactivated mechanisms in cytotoxic T cells,69 following the example of outstanding efficacy of anti-PD1 plus chemotherapy observed in cHL and lung cancer.70-72 A clinical trial sponsored by the National Cancer Institute (United States) evaluating nivolumab in combination with immunochemotherapy for newly diagnosed PMBL is ongoing (NCT04759586), but it is not selecting patients based on their molecular profile. Nonetheless, it seems likely that patients with PDL1low/PDL2low will not benefit from receiving anti-PD1 therapy. We suggest that future prospective trials should investigate anti-PD1 treatment in the subset of PDL1high/PDL2high cases.
In conclusion, we defined a subset of 23% to 30% of patients with PMBL with strong immune privilege based on high gene expression of both the PDL1 and PDL2 genes who have poor outcomes from firstline immunochemotherapy. The RT-MLPSeq assay might serve as a companion diagnostic to identify these patients at high risk of chemotherapy resistance in a timely manner. Other alternatives, including anti-PD1/PDL1 checkpoint inhibitors, need to be investigated as frontline treatments in this specific high-risk population.
Acknowledgments
The authors thank the patients and their families, the reviewers at the LYSA, notably Clémentine Sarkozy and François Lemonnier, and all of the investigators in the LYSA centers. The authors thank the LYSA Pathology group and platform for project management, tumor collection, banking, TMA construction, IHC techniques, and digital slides: Véronique Jaloux, Chantal Brochet, Maryse Baia, Jacqueline Polyte, and Nadine Vailhen. The authors thank the Biological Resource Center (CRB) of the Centre Henri Becquerel for tumor sample banking and nucleic acid extractions: Ludivine Beaussire, Marick Lae, and Sophie Mascre; Julie Libraire, clinical data manager at the Clinical Research Unit, Centre Henri Becquerel; and Doriane Richard, CRA manager, for support in this study. In addition, the authors thank the Centre de Traitement des Données du Cancéropôle Nord-Ouest for data management support; Arthur Dumouchel and Pierrick Gouel for imaging data management in this study; and Lucile Couronné and Maryse Baia for their support in carrying out the FISH analyses. The authors thank the Lymphopath consortium and pathologists of the LYSA centers for sending their samples for the study: J. C. Sabourin (CRB-Centre de Ressources Biologiques-Tumorothèque of Rouen University Hospital), P. Gaulard (Henri Mondor University Hospital, Créteil), V. Meignin (St Louis University Hospital, Paris), C. Glaser (Centre Hospitalier de Versailles), A. Traverse-Glehen (CRB Hospices Civils de Lyon), A. Sudaka-Bahadoran (CRB Centre Antoine Lacassagne, Nice), F. Charlotte (Pitié-Salpetriere University Hospital, Paris), M. C. Copin (Lille University Hospital), L. Martin (Dijon Bourgogne University Hospital), P. Dartigues (Institut Gustave Roussy, Villejuif), C. Chassagne-Clément (Centre Leon Berard, Lyon), M. C. Rousselet (Angers University Hospital), M. C. Chapeau (Centre Hospitalier Départemental de Vendée), M. Patey (Reims University Hospital), A. Moreau (Nantes University Hospital), P. Delvenne (Liège University Hospital), K. Le Dû (Clinique Victor Hugo, Le Mans), P. Tas (Rennes University Hospital), J. Sandrini (CH Le Mans), F. Bibeau (Besançon University Hospital), A. Ledoux-Pilon (Clermont-Ferrand University Hospital), and C. Galant (Cliniques Universitaires Saint-Luc, Bruxelles).
This work was supported by grants from the Ligue Contre le Cancer (Comité de Seine-Maritime, AO_2020), the groupement d'intérêt public Cancéropôle Nord-Ouest (N°2021/01 and N°2021/13), Force Hémato (N°02-2020), and Institut Carnot CALYM (ANR-2020).
Authorship
Contribution: V.C. designed and supervised the study, collected, analyzed, and interpreted the data, and wrote the manuscript; P.S., C.H., E.D., M.P., C.R., L.M., J.P., O.T., A.W., C.A., J.L., and P.G. collected the data; A.T.-G., T.J.M., E.-L.V., and F.D. performed the expert pathologic review and interpreted the data; E.B. performed and interpreted the NGS experiments; V.R. and P.R. performed and interpreted the GEP experiments; S.K. and D.P. performed and interpreted the FISH experiments; F.D., E.-L.V., A.T.-G., and J.B. performed and interpreted the IHC experiments for PDL1, PDL2, and B2M; P.D., S.B., A.B.-R., and D.T. analyzed the metabolic imaging data; P.-J.V. performed the bioinformatics and statistical analysis; E.L. performed the statistical analysis; H.T. analyzed and interpreted the data and edited the manuscript; F.J. designed and supervised the study, analyzed, and interpreted the data, and edited the manuscript; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vincent Camus, Department of Hematology, Centre Henri Becquerel, 1 rue d’Amiens, 76038 Rouen Cedex, France; e-mail: vincent.camus@chb.unicancer.fr.
References
Author notes
∗F.D. and E.-L.V. contributed equally to this study.
†T.J.M. and A.T.-G. contributed equally to this study.
Presented in abstract form (abstract no. 70) at the 17th International Conference on Malignant Lymphoma, Lugano, Switzerland, 16 June 2023.
Data are available on request from the corresponding author, Vincent Camus (vincent.camus@chb.unicancer.fr).
The full-text version of this article contains a data supplement.