Abstract
Bit by bit, over the last few decades, functional genomic tools have been piecing together the molecular puzzle driving tumorigenesis in human patients. Nevertheless, our understanding of the role of several genes and regulatory elements that drive critical cancer-associated physiological processes from disease development to progression to spread is very limited, which significantly affects our ability of applying these insights in the context of improved disease management. The recent advent of clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9)–based technology and its application in cancer genomics has, however, allowed the generation of a wealth of knowledge that has helped decipher several critical questions associated with translational cancer research. Precisely, the high-throughput capability coupled with a high level of technological plasticity associated with the CRISPR-Cas9 screens have expanded our horizons from a mere struggle to appreciate cancer as a genetic disease to observing the integrated genomic/epigenomic network of numerous malignancies and correlating it with our present knowledge of drugging strategies to develop innovative approaches for next-generation precision cancer medicine. Specifically, within blood cancers, current CRISPR screens have specifically focused on improving our understanding of drug resistance mechanisms, disease biology, the development of novel therapeutic approaches, and identifying the molecular mechanisms of current therapies, with an underlying aim of improving disease outcomes. Here, we review the development of the CRISPR-Cas9 genome-editing strategy, explicitly focusing on the recent advances in the CRISPR-Cas9–based screening approaches, its current capabilities, limitations, and future applications in the context of hematological malignancies.
Introduction
The era of functional genomics for cancer research is underscored by 2 groundbreaking feats: human genome sequencing and the discovery of clustered regularly interspaced short palindromic repeats (CRISPR). Although the former achievement is more of a technological derivative, CRISPR-Cas9 is a revolutionary tool for high efficiency, target-specific RNA protein–based editing of the genome with a host of applications in cancer, from cell and gene therapies, diagnostics, biomarker research, basic biology investigation, and several more. Moreover, since the last decade, by complexing hundreds, and sometimes thousands, of individual CRISPR-Cas9 assays in a single experimental setup, in the form of a large-scale multiplexed screen, cancer biologists have developed this technology to be highly scalable so that it can assist in the high-throughput editing and characterization of the complex and heterogenous genetic and epigenetic elements associated with the malignant genome. Although comprehensive short hairpin RNA (shRNA)-based RNA-interference–centered screens or other protein-based genome-editing tools like zinc finger nucleases (ZFNs), meganucleases, and transcription activator–like effector nucleases (TALENs) still have their small pool of loyal followers; the unmatched technological flexibility, target modulation efficacy, and very limited off-target effects offered by CRISPR-Cas9 screening puts it on a pedestal in cancer biology research. Specifically, within blood cancers, the last few years have witnessed a massive turnout of very high impact publications incorporating genome-wide or limited target pool–focused CRISPR-Cas9 screens in virtually every aspect of cancer biology, from the identification of determinants of drug sensitivity, diagnostic and prognostic biomarkers, genetic and epigenetic factors impacting the disease development from preneoplasia to malignant neoplasia, prospective therapeutic targets, molecular mechanisms of novel drug approaches, and encapsulating molecular interplay using in vivo cancer models. With blood cancers commonly serving as the first experimental model for possibly every major therapeutic and technological innovations that are subsequently widely adopted, the data derived from these CRISPR screen–based studies are also expected to have a large impact on shaping high-throughput pan cancer research in the current and future. Keeping this in mind, this review provides an overview of the developmental history of CRISPR-Cas9 technology with particular focus on the current application of the CRISPR screening platform, specifically within hematologic malignancies.
Evolution of CRISPR: from immune regulators to genome engineering tools
History of the CRISPR-Cas system
Although a detailed account of the nearly 20-year history of CRISPR has been reviewed in detail elsewhere,1 the following subsection will provide a brief overview on the developmental history of this technology to establish a foundation to CRISPR-based screening, which remains the focus of this review. The earliest recorded version of the CRISPR system was identified serendipitously in the 1980s in Escherichia coli during an attempt to detect the protein responsible for the isozyme conversion of alkaline phosphatase.2 Within the next decade, the development of high-throughput genome sequencing platforms led to the identification of the repeat elements in several archaeal and bacterial genomes.3 A landmark discovery in the mid-2000s identified the CRISPR protein system as a key component of the adaptive immune system that protects against invading mobile genetic elements by developing memory spacer sequences uniquely derived from the mobile genetic element and inserting them into CRISPR arrays.4-7 Around the same time, a group of genes that were believed to encode DNA repair proteins specific for thermophilic archaea8 were found to associate exclusively with the CRISPR system and protect the prokaryotic cells from viral or plasmid attacks and were consequently designated as cas genes.9,10 Of importance, in 2005, while studying the bacterium Streptococcus thermophilus, Bolotin et al encountered an unusual CRISPR locus that encoded novel cas genes named cas1B, cas5, and cas6.5 Specifically, the cas5 gene was observed to translate into a large protein with a nuclease activity, and over the period of its widespread applicability in gene-editing studies, this gene was renamed as cas9.
A few years later, a group from The Netherlands identified phage-derived spacer sequences that were transcribed into mature CRISPR RNAs (crRNAs), which then served as “guides” for Cas proteins to locate and attack the invading viral nucleic acids, thereby interfering with virus proliferation in E coli.11 In the same year, the CRISPR-Cas system was shown to directly target the phage DNA and confer sequence-directed immunity in the pathogen Staphylococcus epidermidis.12 This DNA-targeting role of the CRISPR-Cas system was corroborated in a subsequent study using in vivo evidence to demonstrate that the S. thermophilus CRISPR-Cas system specifically cleaves plasmid and bacteriophage DNA in a protospacer-specific and orientation-dependent manner.13
In 2011, the Siksnys laboratory expressed the CRISPR-Cas locus from S. thermophilus in E. coli and observed that the organism was capable of plasmid cleavage and resistance.14 Moreover, Cas9 was also established as the only protein required in the CRISPR-Cas9 system, in conjunction with the crRNAs to enable DNA recognition and cleavage; a defining feature of the type II CRISPR/Cas system that made it highly valuable in genome engineering applications.13,14 Indeed, the Siksnys group utilized their heterologous system to characterize the Cas9 protein.15 Specifically, they observed that Cas9 cleaves the DNA strand complementary to the RNA sequence using its HNH domain, whereas the RuvC domain of Cas9 cleaves the antiparallel DNA strand. The binding specificity of the Cas9 protein was determined by a 20-nucleotide sequence preceding the 3-nucleotide protospacer adjacent motif that is complementary to the crRNA. Following a double-strand break (DSB) in the target sequence, the host cell has 2 alternative repair mechanisms: the error-prone nonhomologous end joining mechanism that introduces insertions or deletions, causing a loss-of-function of the target gene, or the homology-directed repair mechanism that incorporates assisted recombination to reconstruct the cleaved DNA. In contrast to the protein-DNA interaction exhibited by ZFNs and TALENs, CRISPR depends on Watson-Crick pairing between crRNA and the target DNA (Figure 1). Consequently, by changing the sequence of the crRNA, the Cas9 protein could be reprogrammed to target any site of choice.
Overview of the role of genome/gene-editing platforms. Genome-editing nucleases (TALEN, ZFN, and CRISPR-Cas9) induce DSBs within the nuclear DNA at specific sites, which can subsequently be repaired using nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) pathways that assist in targeted modification of the DNA. In contrast, gene-editing RNA interference strategies (using shRNA or small interfering RNA [siRNA]) bind to complementary messenger RNA sequences within the cytoplasm and cause its degradation. mRNA, messenger RNA; RISC, RNA-induced silencing complex. Created with www.BioRender.com.
Overview of the role of genome/gene-editing platforms. Genome-editing nucleases (TALEN, ZFN, and CRISPR-Cas9) induce DSBs within the nuclear DNA at specific sites, which can subsequently be repaired using nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) pathways that assist in targeted modification of the DNA. In contrast, gene-editing RNA interference strategies (using shRNA or small interfering RNA [siRNA]) bind to complementary messenger RNA sequences within the cytoplasm and cause its degradation. mRNA, messenger RNA; RISC, RNA-induced silencing complex. Created with www.BioRender.com.
A significant chapter to the CRISPR story was also drafted by the Charpentier group in 2011 when they uncovered the transactivating crRNA (tracrRNA), a transencoded small RNA upstream of the type II CRISPR-Cas9 locus in Streptococcus pyogenes, which was observed to be vital for crRNA maturation by RNase III, a factor critical for the protection of S. pyogenes from the viral invaders.16 In the subsequent year, the Charpentier group collaborated with the Doudna laboratory and reported that the crRNA and tracrRNA could simply be combined to create a single, synthetic RNA chimera, which later evolved as the single-guide RNA (sgRNA) that can induce a sequence-specific Cas9-directed dsDNA cleavage.17 The remarkable contributions of Charpentier and Doudna won them the 2020 Nobel prize in chemistry for their work on CRISPR-Cas9–based genome-editing technology.18 Following the large body of work by the Siksnys, Charpentier, and Doudna laboratories that laid foundation to the role of CRISPR-Cas9 in genomic engineering, subsequent studies by Feng Zhang at the Broad Institute of Massachusetts Institute of Technology and Harvard, the Doudna Laboratory of the University of California, and George Church at Harvard University applied the CRISPR-Cas9 system in eukaryotic cells and showed that the system can be programmed to target multiple genomic loci.19-21
The multifacets of the CRISPR-Cas9 system in genome functionalization
Since the initial implementation of the CRISPR system in mammalian cells, there has been an unprecedented increase in the identification and/or synthetic development of CRISPR-associated molecular components that have been applied to solve specific problems. One of the key proteins in the gene-editing assembly which has undergone significant purpose-driven evolution, is Cas. The most prevalent form of Cas9, the S pyogenes–derived SpCas9, is a highly efficient sgRNA-guided ribonuclease that has been engineered to recognize the canonical protospacer adjacent motif sequence (NGG) as well as its variants (GAT, GAA, and NG).22-24 Other Cas9 orthologues, including SaCas9 (derived from Staphylococcus aureus), NmeCas9 (from Neisseria meningitidis), or CjCas9 (Campylobacter jejuni) have a smaller size relative to SpCas9, consequently rendering them suitable for size-limited delivery vectors such as adeno-associated virus.25-28 Alternative Cas forms such as Cas12a (Cpf1) function independent of a tracrRNA and cleave DNA via a staggered DNA DSB,29 whereas Cas13 (C2c2) cleaves single-stranded RNA instead of DNA and thus is very effective as an antiviral for RNA viruses when investigating viral localization, replication, evolution, and polymorphisms.30-33
Moreover, synthetic Cas variants fused with vital enzyme subunits or whole enzymes have been shown to function effectively in improving target editing for engineering cells and animals for gene discovery. Notable fusion variants include the catalytically dead dCas13b fused with the human RNA–modifying adenine deaminase domain of adenosine deaminase that can induce targeted deamination in RNA,34 catalytically impaired dead Cas9H840A fused to a reverse transcriptase programmed with a prime editing guide RNA that allows for target specification and site-specific editing,35 nuclease-defective Cas9D10A nickase variant fused to a deaminase that results in clean, targeted transition base substitutions without the need for a DSB,36,37 fusion of catalytically inactive Cas9 and FokI nuclease that significantly improves the DNA cleavage specificity as compared with the wild type Cas9,38,39 and programmable DNA-binding domains such as ZFNs or TALENs fused with Cas9 that increases precision and targeting range.40 Apart from Cas enzyme modifications, the use of truncated sgRNAs or partial replacement of RNA nucleotides with DNA bases in the crRNA sequence are potential strategies to significantly reduce unwanted cleavage at off-target sites or cleavage of homology-directed repair–corrected sites, thereby enabling precise genome editing.41,42
With the improvement in the targeting efficiency of the components of the CRISPR-Cas system, its application as a potent genome engineering tool evolved simultaneously. In addition to modifying genomic DNA sequences, research groups around the world started applying CRISPR to regulate target genes. Different CRISPR activators such as dCas9 fused with the transcriptional activation domain VP64 or tripartite transactivation complex VP64-p65-Rta have been used to robustly induce the expression of targeted genes.43-46 In contrast, CRISPR inhibitor systems employing dCas9 fused with Kruppel-associated box transcriptional repressor domain inhibits the transcription of target genes.47,48 Similarly, such synthetic fusion variants of dCas9 have been employed to induce locus-specific epigenome editing including site-specific DNA methylation (dCas9 fused with DNA methyltransferase),49-51 targeted demethylation (dCas9 fused with ten-eleven translocation proteins),52,53 post-translational modification on histone tails (dCas9 fused with LSD1),54 histone deacetylation (dCas9 fused with histone deacetylase),55 and a lot more.
Overall, the historical and ongoing evolution of CRISPR coupled with an array of functional modalities makes this technology very valuable in genomic engineering compared with other players in this field. Protein-based nuclease systems such as ZFN and TALEN suffer from complex design approaches, lower target specificity, and overall low system efficiency. Other strategies such as complementary DNA libraries can only be utilized for gain-of-function whereas small interfering RNA and shRNAs can only cause downregulation of the messenger RNAs (Figure 1). Moreover, the effect of the RNA interference–based screening is prominent for a short time frame posttreatment. Although CRISPR has its own shortcomings, such as off-target effects and the potential long-term impact of Cas9 in target cells,56 it is an actively developing technology, which is rapidly becoming the go-to tool for scientists studying a multitude of biological processes.
CRISPR-Cas9 screening
Apart from the targeted genetic and epigenetic manipulations, the simplistic, yet highly efficient, gene targeting capacity of CRISPR has been exploited to achieve genome-wide screenings using pooled library approaches or arrayed libraries (Figure 2). Such studies typically use lentiviral libraries comprising several hundreds or thousands of sgRNAs targeting the entire genome, a specific family/set of genes, or the noncoding genomic regions, including enhancer elements, promoters, and intergenic regions in a cell population. The wide applicability of CRISPR-Cas9 functional screens puts them at a significant advantage relative to RNA interference–based screening, which can only generate a low-efficiency unilateral outcome of gene expression reduction that is limited to coding genes.
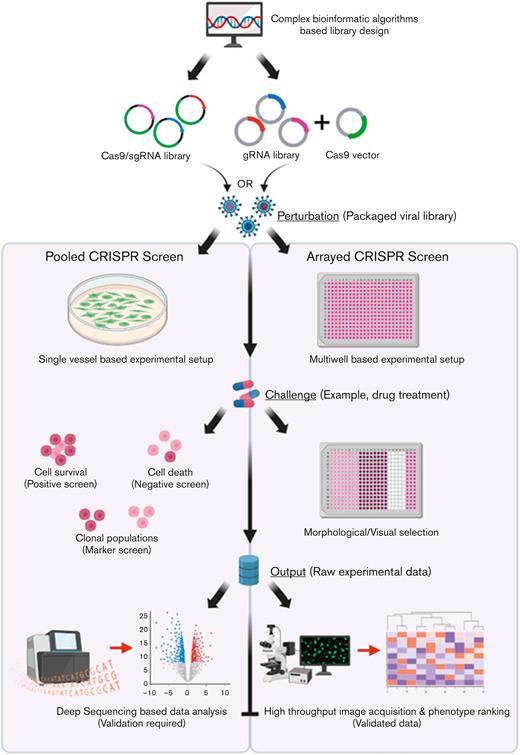
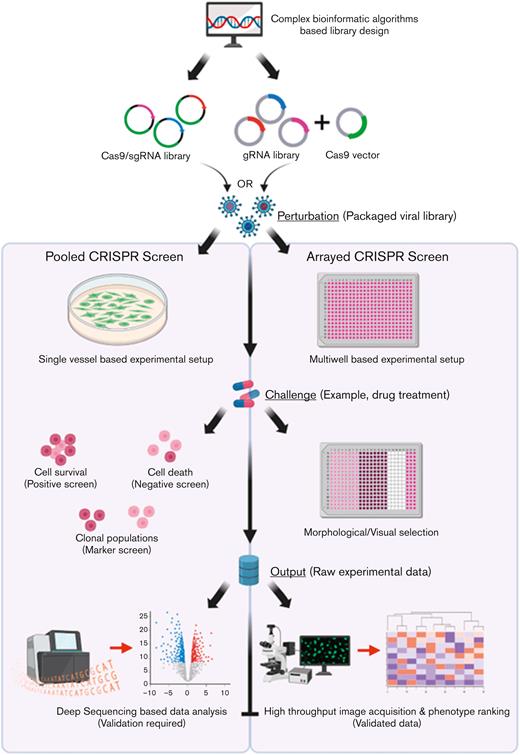
Workflow of pooled vs arrayed CRISPR-Cas9 screening approaches. For either screening strategy, the input pool of sgRNAs and Cas9 is designed using complex bioinformatic algorithms to maximize targeting efficiency and limit off-target effects. In the pooled screening approach, a viral sgRNA library generated from the sgRNA/Cas9 complexes can then be delivered to a single vessel of cells at a low MOI. In contrast, within the arrayed screening format viral libraries are delivered to discrete cell populations grown in multiwell plates to allow for unique representation of individual sgRNAs within each well. Following the administration of a challenge (such as drug treatment), based on the selection strategy adopted (positive, negative, or FACS-associated marker based), several rounds of cell selection may be included within pooled screens to allow for targeted cell enrichment. In contrast, the arrayed screens do not require selective enrichment, rather the developing phenotype within the individual cell populations form the raw data that can be visualized in the final step of the assay through high-throughput image acquisition strategies followed by statistical ranking of the observed phenotypes. In contrast, raw data (genomic DNA from the selected cell populations) must be profiled using deep-sequencing approaches followed by an analysis of depleted or enriched sgRNA population to identify the potential genes that could be associated with potential cancer pathways although downstream validation will be required to confirm the speculative role of the potential genes within the disease. Key steps of convergence between the 2 pathways have been highlighted for both pathways. gRNA, guide RNA. Created with www.BioRender.com.
Workflow of pooled vs arrayed CRISPR-Cas9 screening approaches. For either screening strategy, the input pool of sgRNAs and Cas9 is designed using complex bioinformatic algorithms to maximize targeting efficiency and limit off-target effects. In the pooled screening approach, a viral sgRNA library generated from the sgRNA/Cas9 complexes can then be delivered to a single vessel of cells at a low MOI. In contrast, within the arrayed screening format viral libraries are delivered to discrete cell populations grown in multiwell plates to allow for unique representation of individual sgRNAs within each well. Following the administration of a challenge (such as drug treatment), based on the selection strategy adopted (positive, negative, or FACS-associated marker based), several rounds of cell selection may be included within pooled screens to allow for targeted cell enrichment. In contrast, the arrayed screens do not require selective enrichment, rather the developing phenotype within the individual cell populations form the raw data that can be visualized in the final step of the assay through high-throughput image acquisition strategies followed by statistical ranking of the observed phenotypes. In contrast, raw data (genomic DNA from the selected cell populations) must be profiled using deep-sequencing approaches followed by an analysis of depleted or enriched sgRNA population to identify the potential genes that could be associated with potential cancer pathways although downstream validation will be required to confirm the speculative role of the potential genes within the disease. Key steps of convergence between the 2 pathways have been highlighted for both pathways. gRNA, guide RNA. Created with www.BioRender.com.
Pooled screens
The underlying difference between pooled and arrayed screening formats is the way in which the sgRNA library is delivered to the target cells. In pooled screens, a large “pool” of sgRNA-containing vectors is delivered to the target cells in a single vessel at a low multiplicity of infection (MOI) to limit the uptake of >1 sgRNA by a single cell clone, which could affect the overall phenotype exhibited by the cell. The design of the sgRNA library is critical and is commonly developed using a host of bioinformatic algorithms57-61 with the inclusion of multiple sgRNAs (∼4-8) against a single target to increase the statistical certainty of achieving a hit. Commonly, lentiviruses or retroviruses are utilized for delivering the library of perturbations to the cells, owing to their small size, ability to integrate within the host cell genome, and the availability of established protocols for fine tuning the delivery efficiency. To study the phenotype of interest, a parallel cell pool setup, commonly referred to as the control cells, is generated that does not develop the phenotype shown by the target cells. After several rounds of selection pressure, which could last a few weeks, deep sequencing of the polymerase chain reaction–amplified genomic DNA from the clonal populations (treated vs control) is performed using sgRNA-specific sequencing primers. By mapping the raw reads against the sgRNA library used for the study, the enriched or depleted sgRNAs are identified, which are then directly correlated with the corresponding genic or nongenic targets of individual sgRNAs. The inclusion of multiple sgRNAs against the same target, sgRNAs that target known positive and negative regulators as well as nontargeting scrambled controls, greatly assists in identifying true hits responsible for the development of the phenotype of interest.
Considering that pooled screens have a simplistic working protocol, lower operational costs, and do not require complex robotics for liquid handling, the application of such screens is significantly higher for genome-scale studies.62 Indeed, the first CRISPR-based screens were conducted in a pooled library format and demonstrated the remarkable advantage of CRISPR over small interfering RNA in mammalian cells. One of the earliest studies was from the Zhang laboratory who developed the GEnome-scale Crispr-cas9 KnockOut (GeCKO) library comprising of 64 751 unique sgRNAs targeting 18 080 genes (3-4 sgRNAs per gene) to identify factors critical for cell viability in cancer and pluripotent stem cells.63 At the same time, the Lander laboratory incorporated an equally large library of 73 151 sgRNAs targeting 7114 genes (10 sgRNAs per gene) and performed a screen to identify markers of resistance to the nucleotide analog 6-thioguanine in 2 human cell lines.64 Both studies were similar in several aspects. They both made use of a lentiviral mode of sgRNA delivery to the target cells and designed a screen to allow for positive and negative selection. Although positive selection identifies enriched sgRNAs and is quite commonly applied because of the ease of usage, negative selection identifies depleted sgRNAs and is consequently performed on a large-scale to increase detection sensitivity. An important point of difference between these 2 studies was the way in which Cas9 was delivered to the cells. Whereas the Lander group first generated stably transduced Cas9-expressing cell lines, which were subsequently infected by the lentiviral GeCKO library, the Zhang laboratory simultaneously delivered the 2 CRISPR elements by making use of lentiviral vectors comprising adjacent sgRNA and Cas9 expression cassettes. Classically, a 2-vector system delivering the Cas9 and the sgRNAs separately offers the obvious advantage of a higher transduction efficiency and a lower background noise than the single bulky vector system. Although a lower MOI ensures a better representation of the sgRNA library, no such limitation exists for Cas9; and indeed, a higher level of Cas9 per cell ensures a greater knockout efficiency. In contrast, simultaneous delivery of the Cas9, sgRNAs, and the selection marker reduces the workflow and increases the applicability of this system to any cell type of interest.63 Regardless of the differences in the study design, both studies reported a highly targeted allele modification (nearly 100%) with limited off-target effects.63,64
To demonstrate the applicability of the CRISPR screening model in nonhuman mammalian genomes, Koike-Yusa et al made use of a genome-wide CRISPR screening library to target the protein coding genes in mouse embryonic stem cells for comprehensive loss-of-function screening.65 Using this study design, the authors identified 27 previously known and 4 unknown genes showing resistance to either Clostridium septicum alpha-toxin or 6-thioguanine, thereby demonstrating the high power, unmatched performance, and fantastic utility of this technology within the mice genome.
Although genome-wide screens had been shown as highly effective and informative tools, the challenges associated with managing a bulky library encouraged Zhou and colleagues to make use of a significantly smaller lentiviral library of 869 sgRNAs targeting 291 genes (∼3 sgRNAs per gene) within the human HeLa cell line to identify host genes essential for the intoxication of cells by anthrax and diphtheria toxins.66 The defining feature of this study was the usage of a focused library, referred to by the authors as “knowledge-based screening,” to conduct functional genomics.
Whereas these initial studies were purely viability screens, the experimental setup of pooled screens have since been modified to perform several different screening assays. By using the principles of fluorescence-activated cell sorting (FACS), the CRISPR library–infected cells can be physically separated into phenotypically distinct groups carrying high or low expression of a marker of interest. This approach has been successfully used in screens commonly studying immune cells.67-70 Although this strategy is not only time consuming but also expensive, considering that a significantly large population of cells must be processed by FACS to ensure a genome-scale screen; yet, the incorporation of marker-based cell enrichment strategies like magnetic beads or microfluidics have helped in addressing this shortcoming.71-74
The traditional CRISPR technology combined with a single-cell RNA sequencing is another form of pooled screening assay that has recently been used widely to interrogate gene expression profiles of individual cells.75-79 Although the screen is highly focused, incorporating a limited number of sgRNAs, the amount of information generated from such screens is massive.62 As a result, this approach is well suited for complex heterogenous cell systems such as organoids and primary tissues.74,80,81
Apart from the in vitro models, pooled screening assays have also been applied in vivo to better understand the complex molecular interplay of target cells in the natural biological environment. Indirect in vivo screens are a more common, straightforward, and an effective approach that simply involves (1) the development of a stable cell line infected with the sgRNA library and (2) transplantation of the modified cell line into an in vivo model. The earliest genome-wide in vivo CRISPR screen made use of this approach to transplant a stable sgRNA library–transduced nonmetastatic cell line into a nude mouse, and subsequently study the distant lung metastases formed, by means of deep sequencing to identify enriched sgRNAs that targeted genes responsible for metastasis.82 This study was followed by several such indirect in vivo screens specifically to study disease biology.67,83-88 Although indirect in vivo models have a limited working scope, considering that most of these studies rely on nonautologous transplantation of cells in the mouse biological environment, and hence, to avoid recognition and rejection of the target cells by the host immune system, only immunocompromised models can be used.82 In addition, challenges in surgical accessibility to certain solid tissues, such as brain, make functional engraftment of transplant cells difficult, further limiting the operational scope of indirect in vivo models.89,90 To circumvent these limitations, direct, autochthonous in vivo genome-wide CRISPR screening models have recently been successfully applied in the discovery of several potential targets implicated in a multitude of biological processes and disease.91-96 In principle, the direct in vivo screens in cancer have allowed (1) the tumors being modeled to develop from the target endogenous tissue, (2) the host organism’s immune system to be preserved, and (3) the tumor microenvironment to remain intact, thus ensuring the replication of the true tumor functional biology within the murine environment.97 Regardless of the advantages, current direct in vivo screens lack operational flexibility, have a smaller focused screen, a low screening efficiency, and consequently, limited applicability, all of which should hopefully improve as this form of CRISPR screening evolves.
Arrayed screens
Although pooled screens technically work on a forward genetics approach where a set of perturbations are introduced in the target cell population and based on the phenotype exhibited, the corresponding genotype is predicted; arrayed screens operate on a reverse genetics strategy where a specific gene is altered, and the resulting phenotype is investigated. Specifically, arrayed screening libraries incorporate multiwell plates or an arrayed format of cells that are exposed to sgRNAs targeting a single gene within each well, allowing for a clear genotype-phenotype correlation with no data deconvolution. Consequently, this working format allows the user to apply more library types and library delivery methods. Besides, each reaction well is subjected to a different perturbation and hence, the phenotypic readout from the arrayed screen essentially does not require deep sequencing of the sgRNAs.74 For this reason, focused array screens operating with a few sgRNAs are commonly used as a validation approach for the results generated from large, pooled CRISPR screens.98 Furthermore, because each cell population is exposed to a sgRNA targeting a different gene, the initial delivery of the lentiviral sgRNA vector can be done at a high MOI to ensure a high representation of the sgRNA expression cassette.99 Better still, because the arrayed screens operate on a reverse genetics approach, they do not depend on a biosafety level 2 facility requiring highly integrating lentiviruses99 and can work with nonintegrating adeno-associated vectors,100 plasmid vectors,101 synthetic oligonucleotides,102,103 or ribonucleoproteins.104 Although there are several advantages of arrayed CRISPR screens, this technological format is developing slowly, owing to the requirement of robotics for large-scale screens, low flexibility, and limited application relative to pooled screen approaches. With more research groups making use of CRISPR screening assays for target discoveries, there will be a gradual increase in the dependency toward arrayed CRISPR screens and its reverse genetics approach, which carries a higher biological relevance.
Application of CRISPR screening to investigate the biology of blood cancers
With the advancement of tumor sequencing in the past decade, we now have a significantly better understanding of the catalog of genetic alterations in almost every cancer type. The application of RNA interference screening libraries for the discovery of markers of tumor biology (development, angiogenesis, metastasis, resistance, etc) and potential drug targets allowed the large-scale identification of probable cancer-associated genes, thereby increasing our knowledge and understanding of this disease. In the last few years, the advancement of CRISPR-based screening strategies has complemented and substantially extended the discovery approaches in several solid tumors including lung,82,105,106 breast,107-109 brain,87,94,110 colorectal,86,111 liver,96,112 and renal cancer.113,114 Within these studies, the multitude of CRISPR screens have enabled high-throughput interrogation of driver and passenger genes in varied tumor biologies, including tumor growth, metastasis, tumor microenvironment, synthetic lethal interactions, therapeutic response, and resistance.115
In contrast to the solid cancers, hematologic malignancies have always been “easy systems” to work with. Owing to their liquid form, blood cancers offer a greater ease of access to the malignant cells, and consequently drug delivery; hence, researchers in this field have frequently led the way in adopting novel therapeutic strategies that have then been widely applied in other malignancies.116 From a CRISPR point of view, blood cancer cell lines are one of the simplest yet robust working models for gene editing and expansion and enrichment of manipulated cells. Consequently, CRISPR-based screening approaches have been applied on almost every subtype of blood cancer to maximize basic, translational, and clinical research in this field. Nearly 200 independent publications from multiple research institutes distributed globally have led to remarkable discoveries associated with the identification of drug resistance mechanisms, novel therapeutic targets, disease biology interrogation, as well as for assessing therapeutic mechanisms of candidate drugs in several hematologic malignancies. A detailed summary of these research achievements is presented in the supplemental Table 1. Considering the large number of research outputs and the unique nature of each publication, it would be impossible for us to touch base on all studies owing to a limitation on the length of this review. Nevertheless, in the following sections, we will attempt to identify key research outputs within the 3 major blood cancer subtypes: myeloma, leukemia, and lymphoma, to better illustrate the power for high-throughput process handling, sensitivity for fine tuning target selection, and the flexibility to optimize multiple research approaches of the CRISPR-based screening platform in blood cancers.
CRISPR screens in MM
A major subtype of hematologic malignancies is multiple myeloma (MM) that is characterized initially by malignant plasma cells in the bone marrow, which subsequently spreads to extramedullary sites and peripheral blood as the disease progresses. Plasma cells are terminally differentiated B cells, responsible for secreting antibodies and maintaining the humoral immune response. Consequently, the clinical manifestation of MM includes a significant rise of clonal MM cells in the bone marrow accompanied with ≥1 signs of organ damage as well as hypercalcemia, renal failure, anemia, cytopenia, and immune paralysis.117 Although the developmental stages, monoclonal gammopathy of unknown significance and smoldering myeloma, are premalignant, all of these stages share numerous similar genetic features with MM.117 The development of several generations of new therapies, from traditional chemotherapy to advanced therapeutic strategies such as proteasomal inhibitors and immunomodulatory drugs (IMiDs) to monoclonal antibodies, histone deacetylase inhibitors, and finally autologous stem cell transplantation, in the last few decades has significantly improved the overall outcome of MM.118 Yet, an estimated 176 404 new MM cases and nearly 120 000 deaths were recorded globally in 2020 alone,119 indicating the imminent requirement of novel therapies to better manage this disease.
A key factor responsible for MM progression is the development of therapeutic resistance. Consequently, the majority of CRISPR-based screening approaches in MM have been focused on the identification of critical genes implicated in these resistance pathways. The first study to apply a genome-wide CRISPR-Cas9 screening in hematological malignancies was performed in MM cells, which attempted to identify and delineate mechanisms of proteasomal inhibitor sensitivity.120,121 Procedurally, Hu et al120 and Sheffer et al121 incorporated the GeCKO v2 sgRNA library from the Zhang laboratory in an in vitro MM cell line model (RPMI8226 cell line), which was then treated with clinical anti-MM proteasomal inhibitor drugs such as bortezomib (BTZ) and bromodomain inhibitor JQ1. The surviving cells were subsequently profiled by deep sequencing to identify candidate genes/pathways regulating the resistance events. To obtain a focused set of hits, the authors further performed 2 concurrent CRISPR screens in a larger panel of MM cells lines using a targeted library of sgRNAs against candidate resistance genes. The identified candidate BTZ-resistance genes were observed to play important roles in the pathways associated with regulation of apoptosis, autophagy, chromatin remodeling, heat shock protein expression, aggresome function, etc, which were also found to be frequently mutated, deleted, or downregulated in newly diagnosed MM that showed a shorter time to disease progression after treatment with BTZ.
A subsequent study by Shi et al operating on a very similar working model, applied an unbiased genome-wide CRISPR screening approach and identified the proteasome-regulatory subunit PSMC6 as the most essential gene associated with BTZ resistance.122,123 Indeed, PSMC6 was also reported independently by Hu et al and Sheffer et al and also in a separate shRNA-based study conducting a knock down of the gene in MM cells124 as a prominent factor responsible for BTZ-associated resistance,120,121 not only indicating the importance of this gene as a potential therapeutic target and/or a marker of drug resistance but also providing clear proof of the highly sensitive nature of the genome-editing screening platform. Moreover, the flexibility and the high-throughput environment offered by the discovery model of genome-wide CRISPR-based screening approaches have been instrumental in the validation of previously identified speculative targets, aside from novel candidate identification in a host of cell processes.
Taking advantage of this phenomenon, Bohl et al applied comprehensive CRISPR screens, custom developed using a panel of relapse-associated genes identified from the whole-exome sequencing of patients showing disease relapse after chemotherapy, and uncovered genetic determinants of resistance against proteasomal inhibitors (BTZ), IMiDs (lenalidomide), glucocorticoids (dexamethasone), and cytotoxic chemotherapy (melphalan) in MM.125 Top hits identified included RB1 and CDK2NC (resistance markers for BTZ), cereblon (CRBN), and other CRBN E3 ligase members DDB1 and CUL4B (resistance markers for lenalidomide), structural genes PCDHA5 and ANKMY2 (resistance markers for dexamethasone), and TP53 (resistance marker for melphalan), indicating that the resistance patterns identified were highly dependent on the drug therapy administered and showed limited overlap.
Although more such studies are required to evaluate the impact of these candidate gene alterations on MM treatments, the mere identification of these leads is a positive step toward the development of personalized treatment approaches in this disease. Indeed, genome-scale screens by Sievers et al126 and Liu et al127 report a similar family of genes (ubiquitination associated) being implicated in the therapeutic sensitivity of MM cells toward IMiDs, critically validating the importance of these genes as potential therapeutic targets in MM. Mechanistically, the IMiDs were observed to bind the protease CRBN and activate cullin–RING ubiquitin ligase complex 4 (CRL4)/CRBN E3 ubiquitin ligase to cause the proteasomal degradation of important B-cell–specific transcription factors, Ikaros family zinc finger 1(IKZF1) and Aiolos (IKZF3), and induce MM cytotoxicity.128,129 Specifically, Sievers et al demonstrated a unique role of the ubiquitin donor E2 enzymes UBE2D3 and UBE2G1 in cooperating with CRL4/CRBN E3 ubiquitin ligase for ubiquitinating IKZF3, which consequently impacts the overall activity of IMiD.126 On similar lines, Liu et al highlighted the importance of a constitutive photomorphogenesis 9 signalosome subunit that modulates CRBN expression levels and consequently the sensitivity of IMiDs in MM.127 Using this candidate discovery to their advantage, the authors showed that pretreatment of MM cells with proteasome inhibitors or NEDD8-activating enzyme inhibitors eliminates the impact of the constitutive photomorphogenesis 9 signalosome complex on CRBN, thereby increasing the effectiveness of IMiD therapy. Likewise, several alternative therapeutic strategies have been discovered including the cardioprotective drug dexrazoxane that inhibits DNA topoisomerase IIβ130 and MEK inhibitors,131 which have been observed to overcome IMiD resistance in MM and improve patient survival.
Taken together, CRISPR screens in MM have been highly informative in uncovering vital leads that bear significant potential in therapeutic resistance and the overall disease outcome. However, there have been very limited CRISPR-based screens performed in MM to discover key markers associated with the development of this disease, which is perhaps an area of investigation that should also be focused on to gain further insight in the biology of MM.
CRISPR screens in leukemia
In contrast to MM, leukemias are highly heterogenous hematological malignancies that arise from either lymphocytes or myelocytes, both of which can develop either as a rapidly progressive disease with immature, underperforming leukocytes (acute form), or progress slowly wherein the affected cells are well differentiated yet show subpar functionality (chronic form). Owing to the rapid progress of global research to understand the developmental stages, improved existing cytotoxic, targeted, and immune therapies, and the discovery of novel diagnostic and prognostic biomarkers, management and disease outcome in leukemia have significantly improved in recent years.
Myeloid leukemias
Multiple studies have now shown that myeloid leukemias (acute and chronic) are clonal diseases that develop from a series of genetic and epigenetic transformations in hematopoietic stem cells or precursors, which suppresses the normal hemopoiesis and results in the buildup of abnormal myeloid cells in the marrow and peripheral blood. In accordance, a better understanding of the developmental biology of this disease group would give the research community an upper hand in designing potent therapeutic strategies to improve disease management.
An important phenotypic characteristic of myeloid leukemias (specifically, acute myeloid leukemia [AML]) as well as several other cancers is impaired cellular differentiation and an uncontrolled clonal expansion of myeloid precursor cells. Consequently, a critical area of blood cancer research is the identification of differentiation regulators in myeloid leukemias, which can lead to the development of differentiation therapies that can stimulate the maturation of immature myeloblasts as a mechanism to control cancer progression. That being the case, Wang et al incorporated a novel, integrated cell-surface antigen–based CRISPR screen to identify potential regulators of AML differentiation.132 Experimentally, the authors incorporated a FACS-based pooled CRISPR-Cas9 screening assay of target AML cells, which were then sorted based on the expression of differentiation markers CD14 and CD11B and profiled by deep-sequencing sgRNA enrichment. Although they identified several previously known AML dependencies such as DOT1L, HDAC3, MYB, and PRMT1,133-136 the authors specifically uncovered zinc finger protein 36 ring finger protein like 2 as a critical marker promoting deadenylation and degradation of messenger RNA transcripts that regulate cell differentiation, indicating that therapeutic targeting of RNA processing pathways is an attractive target in AML treatment.
Not surprisingly, a subsequent genome-wide CRISPR screen performed by Wang et al identified several previously uncharacterized genes involved in RNA processing pathways that were identified as essential genes for the proliferation and survival of myeloid leukemia cells, indicating that this pathway is a critical therapeutic target in AML.137 Another genome-scale loss-of-function screen in this direction by Erb et al led to the identification of the YEATS domain protein eleven-nineteen leukemia (ENL) as an epigenetic reader that promotes oncogenic transcription in AML by regulating the activity of RNA polymerase II.138 Around the same time, Wan et al reported that ENL promoted histone lysine acetylation and crotonylation and that consequently inhibiting the association of ENL with the chromatin using a targeted inhibitor is a valid therapeutic strategy alone, or in combination with bromodomain and extraterminal inhibitors, for the treatment of AML.139 Adding value to this information, a genome-wide CRISPR screen by Romine et al identified that regulators of monocytic differentiation and aryl-hydrocarbon signaling define the therapeutic sensitivity of patients with AML to bromodomain and extraterminal inhibitors;140 and hence, the use of key differentiation regulators as druggable targets augments the treatment of patients who are therapy refractory. In consonance with this result, Yan et al performed a differentiation-focused CRISPR screen in AML cells and identified the histone acetyltransferase KAT6A that cooperates with ENL to epigenetically regulate transcriptional elongation and activation of MYC and other AML oncogenes.141 Consequently, KAT6A was identified as a fascinating avenue for therapeutic targeting through monotherapy or combination treatment approaches.
These studies clearly demonstrate the unmatched value offered by the discovery model of CRISPR-based screening. Although downstream target validation by means of critical in vitro and in vivo experiments are essential for further functional and molecular investigation of the predicted targets, the screening model in itself is a powerful tool for fine tuning the target selection process. Indeed, the incorporation of genome-wide or focused CRISPR screens has led to the identification of several such potential therapeutic targets in myeloid leukemia, including but not limited to CTCF boundary,142 interaction of glycoprotein CD43 and Siglec-7 immune receptor,143 de novo purine biosynthesis pathway components,144,145 Rio kinase 2,145 and several more.
Lymphoid leukemias
Developing from the malignant transformation and unregulated proliferation of the lymphoid progenitor cells in bone marrow, peripheral blood, and extramedullary sites, the lymphoid leukemias represent a well-characterized form of hematological malignancies. Yet, the identification of novel regulators of the biology of these leukemias will help in the development of new therapies or improvement to existing therapeutic modalities for controlling and/or managing the developing disease. In line with this, Bertomeu et al performed a high-resolution extended knockout library–based CRISPR screen targeting >19 000 Refseq genes, 20 852 alternatively spliced exons, and 3872 hypothetical genes to delineate the essential genes required for growth, proliferation, and survival in a pre–B-cell lymphocytic leukemia cell line.146 Following a robust data analysis, which included information from multiple screens on different cell lines, the group observed that only a handful of genes form the “essentiality signature” within cell lines belonging to a similar disease pool. In principle, this indicates that only a small pool of genes is critical for maintaining cellular fitness within the cancer cell lines, whereas many of them would reflect the synthetic lethal interactions that correlate with the unique spectrum of cancer-associated mutations.146 From a therapeutic point of view, the identification of a disease-specific gene essentiality signature within the human proteome using such an extended knockout library provides a clear insight into the biological mechanisms and phenotypic complexity of the disease, thereby allowing the identification of critical time points as well as novel targets in the cancer developmental process that maximize treatment efficacy.
A similar large-scale genome-wide screen was performed to dissect the relapse-related mutation spectrum and genetic determinants of drug resistance in acute lymphoblastic leukemia (ALL).147 ALL is the most common leukemia observed within the pediatric population, although adults with ALL commonly show a poor disease outcome with therapeutic resistance being one of the common determinants of disease prognosis. Consequently, identification of potential resistance-associated gene targets could assist in the development of unique drugging strategies for the treatment of relapse or refractory disease. By functionally exploring gene-drug interactions using the CRISPR screen, the authors identified recurrent somatic mutations in prominent cancer drivers and tumor suppressors including KRAS, MYC, NRAS, JAK2, JAK3, TP53, CREBBP, etc, along with the presence of branched clonal evolution, as well as several candidate genes driving drug resistance events in ALL.
Indeed, such forms of high-throughput global screens uncover a plethora of data related to the biological course of the disease, providing valuable information for the identification of novel therapeutic avenues and disease management strategies. On these lines, Autry et al conducted an integrative multiomics study in addition to a genome-wide CRISPR screen to identify genetic and epigenetic determinants of glucocorticoid resistance in ALL.148 This approach identified numerous previously known resistance genes and pathways, and the authors discovered a novel drug resistance gene cadherin EGF LAG seven-pass G-type receptor 2 (CELSR2). Although modulation of the expression of CELSR2 was shown to be effective in restoring the sensitivity of leukemia cells to glucocorticoid therapy, the authors also observed that a synergistic combination of prednisolone and venetoclax was highly effective in mitigating drug resistance in vivo. A similar study in this direction was performed by Sasaki et al, who incorporated genome-wide CRISPR-Cas9 dropout screens with an aim of identifying the most potent chemotherapeutic regimen for the treatment of Philadelphia chromosome–like ALL subtype harboring IgH-CRLF2 rearrangement.149 The authors demonstrated that even as Ras was observed to be an essential regulator for cell fitness, administering a combination therapy of gilteritinib (a pan-tyrosine kinase inhibitor), and trametinib (a MEK1/2 inhibitor) killed the ALL-IgH-CRLF2 cells regardless of the Ras mutational status.
Although CRISPR screens have been instrumental in the identification of potential therapeutic targets and markers of drug resistance, the flexibility of this screening approach has also led to the improvement in existing therapeutic approaches. One of the novel immunotherapeutic approaches that has garnered significant attention in recent times is the chimeric antigen receptor (CAR) T-cell therapy. Although the potency of CAR T-cell therapy is extremely high, to date its effectiveness has only been realized in hematological malignancies. So far, only 6 CAR T-cell therapies have been approved by the US Food and Drug Administration and are only available for the treatment of leukemia, lymphoma, or MM. Although the therapies have been effective, a better understanding of the mechanisms affecting CAR T-cell cytotoxicity will reduce therapeutic resistance and disease relapse. In this context, Dufva et al performed an integrated drug profiling of >500 compounds combined with a genome-scale CRISPR-Cas9 loss-of-function screen and identified that SMAC mimetics, which function as apoptotic modulators, can also sensitize the cancer cells for CAR T-cell cytotoxicity.150 Moreover, the death receptor signaling pathway was identified as a vital regulator of the effectiveness of CAR T-cell therapy, and hence, presented itself as a novel therapeutic target for enhancing the impact of cancer immunotherapy. In another recent study, Yan et al applied an unbiased genome-wide CRISPR-Cas9 screen and identified a B-cell lymphoma 2 family protein, NOXA, as a master regulator of CD19/20 CAR T-cell therapy in B-cell malignancies.151 Although the restoration of the cellular levels of NOXA increased the sensitivity of the cancer cells to CAR T-cell therapy, the authors identified a potential role of NOXA as a prognostic biomarker and a novel therapeutic target in B-cell malignancies. Considering the limited therapeutic application of CAR T cells and the recently developed CAR natural killer cell therapies in liquid and solid cancers, such unbiased CRISPR-based screens assist in the identification of prospective novel markers of drug sensitivity, which can then be cotargeted to broaden the treatment potential of the immunomodulatory therapies.
CRISPR screens in lymphoma
The third major subtype of hematological malignancies is lymphoma that comprises a group of heterogenous lymphoid malignancies with differences in clinical behavior and treatment responses.152 Although several subgroups exist, lymphomas are classically subdivided as Hodgkin, if the biopsies contain large, abnormal lymphocytes, referred as Reed Sternberg cells, or non-Hodgkin, in cases where the biopsies are devoid of Reed Sternberg cells. Nevertheless, lymphomas are the most commonly occurring form of blood cancers with >600 000 new lymphoma cases and nearly 280 000 identified globally in 2020 alone,119 warranting critical investigations from the research community to identify the causative factors and potential therapeutic targets for better disease management.
Hodgkin lymphoma
The Hodgkin lymphoma (HL) represents a rare B-cell lymphoid neoplasm. Even though advancement of therapeutic strategies in the past few decades has led to a significant improvement in patient survival rates, a substantial proportion of patients with HL show relapse or resistance to therapy.153 A better understanding of the potential molecular determinants of poor prognosis using high-throughput CRISPR-based screening approaches has assisted in the development of novel therapeutic approaches. One of the notable studies in this direction is by Wei et al who applied a focused CRISPR screen using sgRNA library targeting ubiquitin regulators to uncover drivers of resistance toward brentuximab vedotin (BV)–based treatment.154 BV is an anti-CD30 antibody drug conjugate, US Food and Drug Administration approved for the treatment of advanced-stage HL that functions by delivering the conjugate monomethyl auristatin to CD30-expressing hematopoietic malignant cells to induce cytotoxicity.155 Owing to the established importance of the ubiquitin pathway in modulating drug responses, the authors specifically studied this network and identified the ubiquitin-modifying enzymes A20 and RBX1 as well as NF-κB pathway genes and the ABC transporter gene ABCB1 as key factors regulating BV treatment sensitivity in vitro and in vivo.154
A follow-up study by Song et al profiled the mechanistic role of A20 using a similar ubiquitin regulator–focused CRISPR-Cas9 screen,156 and discovered the linear ubiquitin chain assembly complex (LUBAC) as a critical regulator of A20 in HL. Although LUBAC was observed to drive the NF-κB signaling and influence the ubiquitination of A20, the authors performed a second focused screen to identify regulators of the NF-κB pathway leading to the identification of the MAPK kinase kinase family member transforming growth factor β–activated kinase 1 (TAK1) as a major mediator of oncogenic NF-κB signaling in LUBAC-dependent HL cells. Mechanistically, increased activity of LUBAC promotes Tak1-regulated IKK/NF-κB signaling and linear ubiquitination of the NF-κB essential modulator in A20 mutant HL cells, which results in prosurvival signaling and immunosuppression. Consequently, clinical targeting of Tak1 kinase in HL alone or in combination with potential LUBAC inhibitors were determined as an attractive therapeutic strategy for the treatment of A20-mutant HL.
Non-Hodgkin lymphoma
In contrast to HL, non-Hodgkin lymphomas (NHLs) constitute ∼90% of all lymphomas, of which nearly 85% to 90% are derived from B cells, and the rest arise from T cells and natural killer cells. Consequently, the gene expression profiles of all NHL subtypes (>50) reflect their cell of origin, which nonetheless is also greatly influenced by recurrent genetic, epigenetic, and other molecular changes.157 To decipher the genomic makeup of NHL, Reddy et al158 conducted an integrative analysis of whole-exome and transcriptome sequencing of >1000 patients with diffuse large B-cell lymphoma (DLBCL, a subtype of NHL) and identified ∼150 oncogenic drivers. These hits were then functionally validated using an unbiased CRISPR screen in DLBCL cell lines to specifically identify genes essential for cell growth.158 Interestingly, the authors observed that a prognostic model developed using the shortlisted genetic perturbations was significantly more predictive of the sensitivity to current therapies in DLBCL relative to established standards, thus demonstrating the substantial benefit of applying high-throughput genomic screens for identifying patterns of disease development, which is not well reflected with small-scale technologies. Later, the same research group applied the integrated analysis strategy on Burkitt lymphoma (BL), another subtype of NHL, and identified 72 oncogenic drivers, which were later functionally annotated using a CRISPR screen in BL cell lines.159 Among others, the technology platform led to the identification of several candidates including IGLL5, BACH2, SIN3A, DNMT1, as well as type 1 Epstein-Barr virus (EBV) infection, as frequently mutated data points in BL.
The role of EBV in the pathogenesis of lymphomas such as BL, HL, as well as epithelial malignancies such as nasopharyngeal cancer, gastric cancer, and breast cancer is well established.160 In B cells, EBV establishes a latent infection by encoding the EBV nuclear antigens that convert the primary B cells into lymphoblastoid cell lines (LCLs). Although we have a basic understanding of this process, the key players responsible for the establishment of LCL are not well established. Consequently, Ma et al performed a genome-wide CRISPR screen on a LCL and an EBV+ BL cell line to investigate EBV-transformed B-cell dependency factors.161 The authors unearthed several critical regulatory pathways within the LCLs and the EBV+ BL including the phosphatidylinositol 3-kinase/Akt signaling, Myc regulation, and tumor necrosis factor α pathway, which were observed to be regulated by the EBV-oncoproteins, and hence represent potential targets for therapeutic intervention. A follow-up study by the same group in 2020 employed a genome-wide CRISPR screen in EBV+ BL cells and identified Myc as the key factor regulating the activation of the lytic program of EBV in B cells.162 Furthermore, in a separate study published later, the authors performed another genome-scale loss-of-function CRISPR screen to characterize the epigenetic factors responsible for the maintenance of latency program in BL cells.163 The researchers specifically identified critical methylation- and ubiquitination-associated genes including DNMT1, UHRF1, and PRC1 that were observed to be crucial for latency maintenance and represent important therapeutic vulnerabilities for controlling EBV oncoprotein activation and function.
In addition to the studies making use of CRISPR screens to interrogate the disease biology, several groups have incorporated this technology to investigate the molecular vulnerabilities of existing therapeutic approaches in NHL, with an underlying aim of improving treatment efficacy. R-CHOP (rituximab, cyclophosphamide, doxorubicin hydrochloride, vincristine, and prednisone) is an immunotherapy regimen for the treatment of DLBCL. Although the addition of rituximab improves response rate,164,165 ∼35% of patients show relapse.166 Thomsen et al made use of a lentiviral genome-wide CRISPR screening library to look for genes regulating the sensitivity of rituximab in DLBCL and identified B-cell receptor signaling components, B-cell linker protein (BLNK), and Bruton tyrosine kinase (BTK) as candidate genes regulating rituximab-induced apoptosis in DLBCL.167 In another study, Tong et al performed a genome-wide sensitizing CRISPR screen with the EZH2 inhibitor tazemetostat, and identified IKZF1 as the prime candidate modulating the response of DLBCL cells to tazemetostat.168
Although current therapeutic modalities function effectively as first-line treatment in NHL, several patients exhibit resistance or relapse, necessitating novel and effective therapeutic strategies. Gayle et al identified the drug apilimod as a powerful cytotoxic agent in B-cell NHL (B-NHL) through a unique clinical-stage compound library screening.169 Mechanistically, the authors demonstrated that apilimod has a high specificity to the phosphatidylinositol-3-phosphate 5-kinase and has selective cytotoxicity in B-NHL in comparison with control cells. In addition, using a genome-wide screen, Gayle et al identified key lysosome-related genes that regulated the sensitivity of B-NHL cells to apilimod therapy. Based on the exciting outcome of this study, the authors initiated a multicenter trial to assess the therapeutic efficacy of apilimod in patients with B-NHL.170 In a similar story, Thieme et al evaluated the candidate genes impacting the susceptibility of a DLBCL cell line to treatment with a dual BTK/spleen-associated tyrosine kinase inhibitor drug CG-806.171 The drug is already part of multiple clinical trials (#NCT03893682 and #NCT04477291) for the treatment of lymphoid leukemia, NHL, and myeloid malignancies. Among others, the authors discovered that the Bcl-2 network is the key sensitizer for CG-806 and represents a potential opportunity for combined synergistic targeting of Bcl-2 with CG-806 in lymphoid malignancies. These studies are crucial models demonstrating the strong potential of CRISPR-based screens for improving current and prospective therapeutic regimens within blood cancers (Table 1). Soon, combining drug panels with focused CRISPR screens will hold the key in a thoroughly validated selection of therapy or therapeutic combinations that have a strong target-driven activity and a significantly lower chance of background noise in terms of off-target effects, toxicity, development of resistance, etc.
From kickstarting to accelerating: the future of CRISPR-Cas9 screens in blood cancers
Although it has many advantages, the current level of this screening tool can still be classified as “novice,” because many factors must be realized to improve its widespread application. From a technology point of view, the usage of nonintegrative viral libraries is one of the key requirements to reduce dependencies on a high biosafety-level working environment. Furthermore, even as pooled CRISPR screens offer operative flexibility, they are complex setups requiring a high capital cell number, long experimental screening terms, dependencies on high-throughput genome reading tools like next-generation sequencing, and the requirement of a downstream validation component to filter the output of the screen. In contrast, arrayed screens require a complex upfront setup of specialized laboratory equipment, automation, and although the system produces a validated pool of targets, it lacks flexibility and overall system efficiency. Some of these issues are, however, based on our current lack of understanding of the technology. For example, making use of limited target-focused screens can quickly assist in the fine filtering of our selections from the next-generation–based sequencing readouts in pooled screens. Such technological improvements will not only improve the overall efficiency of the CRISPR-Cas9–based screening tool, rather increase its applicability in a host of problem-solving fronts in blood and other cancers.
Another technological problem that requires significant attention is the off-target effects of large-scale base editing. Unintended base cleavage and mutagenesis within random sequences that may show a high similarity with the targeted DNA sequence may result in an undesirable phenotypic output generating potential false-positive or false-negative targets that may require unnecessary validations. Although a large number of the recent screens in blood cancers make use of prevalidated sgRNA sequences or utilize complex algorithms such as deepcrispr, GuideScan, CRISPR design website, CFD, and E-crisp that can potentially identify and discard sgRNAs with predicted off-target effects at the level of library design or after screening. Consequently, several such studies have demonstrated a low off-target effect of the screening library.173-175 Although gene screens are more efficiently managed owing to the availability of a larger working window, recently Tycko et al identified that noncoding screens incorporate lower specificity sgRNAs owing to the presence of a smaller targetable window.176 Using a leukemia cell line, the authors incorporated a genome-wide noncoding screen for essential CTCF sites in chromatin loop anchors and identified that off-target activity caused most of the fitness effects, even after filtering the sgRNAs with no perfect or 1-mismatch off-target sites. Even if discarding these nonspecific sgRNAs is a potential solution for a clean screen, it compromises the overall targeting ability of the screening tool.177 Nonetheless, there is a large-scale effort to comprehensively map genomic features and a concurrent improvement in targeting efficiency through improvements in sgRNA design, incorporation of Cas variants, library delivery methods, and variations in base editors that could help in the overall improvement of the on-target screening efficiency of CRISPR.
From a functional point of view, within hematological malignancies we have identified only a small percentage of vulnerabilities using the current scale of CRISPR screening approaches, although it may seem otherwise. Although the major myelomas, leukemias, and lymphomas have been explored significantly owing to the ready availability of a wide variety of cell lines, clinical specimens, and other model systems, several underrepresented subtypes such as myeloproliferative neoplasms, myelodysplastic syndromes, and a few more subtypes of the major blood cancers lack significant exploration using the current CRISPR screening data sets. Making use of patient-derived organoids or in vivo animal models could offer a better starting point for a comprehensive profile of multiple subtypes of blood cancers. Additionally, though using direct in vivo CRISPR screens might have its own set of operative difficulties, such screens are of a higher biological value compared against any high-throughput in vitro screens, which fail to consider the impact of the tumor microenvironment and the host immune responses against the developing disease. For example, an in vivo genome-wide CRISPR screen in a mouse model of blast-crisis chronic myeloid leukemia with >90% of undifferentiated leukemia cells uncovered the double-stranded RNA binding protein Staufen 2 as a novel dependency within this disease.88 However, more such in vivo screens are required to better identify potential vulnerabilities in blood cancers.
Moving beyond standard CRISPR screens, single-cell technologies such as perturb sequencing also provide opportunities for screening heterogenous cell populations using hematological cancer phenotypes that extend beyond cell viability.178,179 In addition, improvement in the methods to capture the epigenome after CRISPR perturbations is required because the current tools lack high throughput. Recently a CRISPR-based single-cell combinatorial indexing assay for transposase-accessible chromatin (CRISPR-sciATAC) was performed in nearly 30 000 single AML cells by targeting 105 chromatin-associated genes and the resultant data allowed the identification of the role of chromatin remodelers in the context of disease development.79 Moreover, the authors observed that although the CRISPR-sciATAC technology offers a comparable output to published sciATAC data sets, it incorporates an additional factor of guide RNA capture while maintaining its high-throughput functionality and lower operational costs and time requirement as compared with counterpart technologies such as Perturb-ATAC.180
Finally, a critical application within blood cancers (as well as solid cancers) is the identification of therapeutic strategies or druggable targets that have a potent effect on disease treatment. Although, an important factor to consider here is that dismantling gene functions using CRISPR-based knockout screens is not equivalent to it being fine modulated using small molecules.178 Consequently, an overall improvement in the CRISPR screening technology is required to better control for the operational mechanistic of the small molecule. In this context, drug-CRISPR combination screens could assist in the identification of potential therapy-induced vulnerabilities in blood cancers. In line with this, SMAC mimetics were identified as potential sensitizers to anti-CD19 CAR T-cell therapy in B-cell ALL and DLBCL using one such drug-CRISPR screen.151 Taken in combination with resourceful databases such as the Cancer Dependency Map, such combination or focused screens will help in making valuable discoveries related to therapeutic vulnerabilities in blood cancers.
Conclusion
The technology of CRISPR-Cas9 was not developed overnight, although the momentum with which this technology has advanced in the past decade may make it seem as if it had. Whereas significant scientific research has gone into simply understanding the operating manual of this tool, modification of the prokaryotic immune component into a high-throughput genome screening tool with a plethora of applications in the field of functional cancer biology, has taken a lot of determination. Within blood cancers, CRISPR screens are being applied by a large number of laboratories across geographies in understanding early disease development, identifying factors responsible for disease progression and key elements responsible for the development of drug resistance, the selection of prospective novel therapeutic targets, strategies, and combinations with the best possible outcome, and much more. Although a wide zone of applicability of CRISPR-based screens have already been explored in blood cancers, one area that deserves more attention is the effectiveness and the therapeutic mechanisms of drugs that have recently been approved or are under trial. Hematologic malignancies are often pivotal in the development and application of innovative therapeutic strategies. Although new drug candidates are being advanced with one specific target in mind, it would be more beneficial to explore potential determinants of drug sensitivity through CRISPR screening–based discovery approaches to identify probable solutions that would ensure that the maximum efficacy of the drug is served from (1) recognizing combination therapies that can synergize and enhance the effectiveness of the drug, to (2) locating potential biomarkers that can help prestratify patients into potential drug responder and nonresponder groups with high sensitivity and specificity, and/or (3) prerecognizing the correct disease subtype or patient group that will derive the highest response rate upon treatment with the drug candidate. Although there have been a few such studies in this direction (summarized in Table 1), from large-scale development of novel therapeutic approaches from small molecules, to bispecifics, to CARs, the use of high-throughput CRISPR screening–based deep discovery approaches will add immense value to the clinical application of drugging strategies in blood cancers. Unsurprisingly, with the increasing relevance of CRISPR-Cas9 screens, the technology will soon progress to function as the protagonist in the field of hematological cancer modeling and functional genomics for the identification of next-generation targets for precision medicine.
Authorship
Contribution: D.N.I. and H.C. conceptualized the study; D.N.I. wrote the original draft of the manuscript; A.D.S. critically reviewed and edited the manuscript; and H.C. supervised the study and reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hong Chang, Laboratory Medicine Program, Toronto General Hospital, University Health Network, University of Toronto, 200 Elizabeth St, Toronto, ON M5G 2C4, Canada; e-mail: hong.chang@uhn.ca.
References
Author notes
The full-text version of this article contains a data supplement.

![Overview of the role of genome/gene-editing platforms. Genome-editing nucleases (TALEN, ZFN, and CRISPR-Cas9) induce DSBs within the nuclear DNA at specific sites, which can subsequently be repaired using nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) pathways that assist in targeted modification of the DNA. In contrast, gene-editing RNA interference strategies (using shRNA or small interfering RNA [siRNA]) bind to complementary messenger RNA sequences within the cytoplasm and cause its degradation. mRNA, messenger RNA; RISC, RNA-induced silencing complex. Created with www.BioRender.com.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/10/10.1182_bloodadvances.2022008966/1/m_blooda_adv-2022-008966-gr1.jpeg?Expires=1769357083&Signature=Cd7ttBQaEIdN6zjO8Hw2dcVs5Zh8jo4Yc-WrCMx1eYyiROhq6jHb8sprO6ZLxWkIfsCzj384L6pZ91XV1Z1giS0BfGtOCkyq75TqaECXJ21ErmJ8cNlXJF0WF2jw7gxMhh0-XrKb-fOgjgku3oNpWNsnvvASmbcAofraNxIJUY1Gi~VdJTGXlxK5lJgH0TUoho37VtLfPDEr9gP8i3MfB1vLR1~C~sESQpbiwVkTBKWZXwUqZUwVF-oPsWpd0EyeTFkp7gImPD2cs-jUIORSZUaneDjl05trxUS79AkJ0btTet-NAomX30h-oMUTYJCSWFznz36KTj2xgIKmOeT2Vg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Overview of the role of genome/gene-editing platforms. Genome-editing nucleases (TALEN, ZFN, and CRISPR-Cas9) induce DSBs within the nuclear DNA at specific sites, which can subsequently be repaired using nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) pathways that assist in targeted modification of the DNA. In contrast, gene-editing RNA interference strategies (using shRNA or small interfering RNA [siRNA]) bind to complementary messenger RNA sequences within the cytoplasm and cause its degradation. mRNA, messenger RNA; RISC, RNA-induced silencing complex. Created with www.BioRender.com.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/10/10.1182_bloodadvances.2022008966/1/m_blooda_adv-2022-008966-gr1.jpeg?Expires=1769743982&Signature=blh~enNBopmLJ6g86oOzbvuDTGOvp1yDCRC5RQ1y3kxRdNvLu4i~yF15IRf9WfxcYK9t-Uokk3AMEtxns~8jqD~XDrLdFqNp1m2kA3ioIolnpEF0RNVApYovzdiiYPS55H1BtsOQRBC~z-uKa8SYF89C0e7egFNb6qXO6y9-0b2jVhwkO4QzyDAUNo6uIkuFSbAhHLGFYYCMMK8vTir1PJAH6YqIVohWdmx148MXumwsXbYYGc2E2qGFRuMVq66-pzgVY6NVnt0BREUodKVzHEeD3OL19AUliUbDneVYYb8ww7E6tbx6XWi1fEddKhCLn80HH9TIZ6L0UpSrPfaVFw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)