Key Points
Subtypes of follicular lymphoma are identified by model-based clustering of genetic mutations in a large (n = 548) population-based cohort.
These subtypes imply distinct mechanistic subsets of follicular lymphoma with implications for patient risk and treatment opportunities.
Abstract
Follicular lymphoma (FL) is morphologically and clinically diverse, with mutations in epigenetic regulators alongside t(14;18) identified as disease-initiating events. Identification of additional mutational entities confirms this cancer’s heterogeneity, but whether mutational data can be resolved into mechanistically distinct subsets remains an open question. Targeted sequencing was applied to an unselected population-based FL cohort (n = 548) with full clinical follow-up (n = 538), which included 96 diffuse large B-cell lymphoma (DLBCL) transformations. We investigated whether molecular subclusters of FL can be identified and whether mutational data provide predictive information relating to transformation. DNA extracted from FL samples was sequenced with a 293-gene panel representing genes frequently mutated in DLBCL and FL. Three clusters were resolved using mutational data alone, independent of translocation status: FL_aSHM, with high burden of aberrant somatic hypermutation (aSHM) targets; FL_STAT6, with high STAT6 & CREBBP mutation and low aSHM; and FL_Com, with the absence of features of other subtypes and enriched KMT2D mutation. Analysis of mutation signatures demonstrated differential enrichment of predicted mutation signatures between subgroups and a dominant preference in the FL_aSHM subgroup for G(C>T)T and G(C>T)C transitions consistent with previously defined aSHM-like patterns. Of transformed cases with paired samples, 17 of 26 had evidence of branching evolution. Poorer overall survival (OS) in the aSHM group (P = .04) was associated with older age; however, overall tumor genetics provided limited information to predict individual patient risk. Our approach identifies 3 molecular subclusters of FL linked to differences in underlying mechanistic pathways. These clusters, which may be further resolved by the inclusion of translocation status and wider mutation profiles, have implications for understanding pathogenesis as well as improving treatment strategies in the future.
Introduction
The molecular pathogenesis of follicular lymphoma (FL) is closely linked to that of diffuse large B-cell lymphoma (DLBCL) and the physiological germinal center (GC) reaction.1 Key factors connecting these malignancies to GC biology include BCL2 and BCL6 gene translocations, deregulating apoptotic and transcriptional control, along with aberrant somatic hypermutation (aSHM).2-8 The latter provides a mechanistic link between aberrant targeting of enhanced mutagenesis that underpins antibody affinity maturation and the increased rate of B-cell neoplasia associated with the GC reaction. Targets of aSHM have been identified in observational and experimental contexts.8,9 Mutation profiling has detected additional pathways deregulated in FL; recurrent mutations in epigenetic regulators KMT2D (MLL2) and CREBBP have identified these as candidate early initiating FL pathogenic events.10-12,EZH2 has emerged as a promising therapeutic target following the identification of mutations targeting this histone methylase in FL and studies establishing the importance of this epigenetic regulator in GC biology.13 Beyond epigenetic regulators, recurrent mutations have been detected in genes on various pathways, including cytokine/STAT activation and the B-cell receptor.12,14-16 In DLBCL, mutational heterogeneity resolves into recurrent patterns, which have been independently identified by several groups.17-22 While existing data suggest less mutational heterogeneity in FL than DLBCL, FL is not a homogenous entity; whether recurrent patterns of cooccurring mutations allow distinct subtypes of FL to be resolved remains an open question.
In contrast to the aggressive but potentially curable DLBCL, FL is by nature indolent and generally incurable. Seminal studies have addressed the question of FL progression.12,15 Aggressive disease may remain morphologically consistent with FL or undergo transformation to a pattern that meets the diagnostic criteria for DLBCL. Morphological transformation to DLBCL is linked to branching clonal evolution and emergence of mutations and subclones that are rare or undetectable at diagnosis, while progression without transformation may be linked to the persistence of subclones that are detectably present from diagnosis.12,23
Predicting patients with FL at high risk of early disease progression and/or transformation is clinically important. For DLBCL, recurrent patterns of cooccurring mutations can be used to identify risk groups and suggest novel treatment strategies.18-21 For FL, a molecular risk predictor, the m7-FLIPI,24 which integrates mutational with conventional risk factors, has been derived from clinical trials; however, the extent to which patterns of mutation may inform outcome in a wider patient population remains uncertain.25
Using targeted sequencing to classify DLBCL diagnostic samples from our large “real-world” population-based cohort, we recently reported mutational profiling findings that both confirmed and extended those of others, identifying 5 robust DLBCL subclusters.20 Here, we extend this approach to 548 patients with FL with the aim of assessing whether comparable subclusters can be defined in FL using the same panel, sequencing and assessing surplus material archived at the time of diagnosis for somatic mutations against a pan-hematological malignancy panel of 293 genes. Our analysis, which identifies 3 subsets of FL based on somatic mutational patterns, has implications for pathogenesis, risk stratification, and future treatment strategies.
Materials and methods
Patients and procedures
Data are from the United Kingdom’s population-based Haematological Malignancy Research Network (HMRN) (https://www.hmrn.org). Initiated in 2004, diagnoses within HMRN are made and coded by clinical specialists at an integrated hematopathology laboratory, the Haematological Malignancy Diagnostic Service (https://www.hmds.info). HMRN’s methods and ethical approvals (REC 04/Q1205/69; REC 14/WS/0098) are detailed elsewhere.26-28 The study was conducted in accordance with the Declaration of Helsinki. In brief, covering 14 hospitals and tracking all patients with hematological malignancies (∼2500 per year) through clinical and national administrative systems (mortality and morbidity), HMRN’s patient cohort operates under a legal basis that permits full treatment and outcome data to be collected from clinical records without explicit consent (PIAG 1-05 9h/2007).26
The source cohort for the present report comprised 852 subjects newly diagnosed with FL (ICD-O3 = 9690/3) between 1 September 2004 and 31 August 2012. Of these, 548 (64.3%) had suitable diagnostic material for genetic analysis; 538 (98.2%) had their medical records fully abstracted (supplemental Figure 1). All patients were followed up for progression and death until 31 December 2018.
DNA sequencing
DNA was extracted from surplus formalin-fixed paraffin-embedded (FFPE) material archived at the time of diagnostic biopsy using Qiagen QIAamp DNA FFPE tissue kits (Cat no. 51306). For each, 50-200 ng of genomic DNA was sheared using a LE220 focused ultrasonicator (Covaris) to 100-200 bp fragments. Indexed libraries were generated using a modified SureSelect XT protocol (Agilent Technologies), pooled (16-plex), and captured with 120 nt biotinylated RNA baits (Agilent Technologies) covering 293 genes and associated extended genomic regions (supplemental Table 1) using the SureDesign interface (Agilent Technologies) on default parameters. Capture libraries were quantified, assessed for size distribution and quality, and sequenced (Illumina HiSeq 2500) using 75-base paired-end sequencing. The average read-depth across all samples was 500× reads-per-base. For full details of sequencing, variant calling, and annotation, see supplemental methods section 2.1. Single nucleotide variants and copy number variants are detailed in supplemental Tables 2 and 3.
Mutation signature analysis
All variant calls for each sample were analyzed with Signal (https://signal.mutationalsignatures.com/)29 using default parameters. The nucleotide exchange patterns in the triplet context, along with contributing mutational signature estimates, were generated with default parameters (Bootstraps: 100; Threshold: 5; P value = .05) for either all mutations across defined sets of genes linked to mutational clusters or for individual cases.
Statistical analysis
To identify subclusters defined by genetic features, binary mutation data were modeled as a finite mixture of Bernoulli distributions, with the number of clusters selected using the Bayesian Information Criterion (BIC), while the Akaike Information Criterion (AIC) was applied to identify the finer structure.20 As the signal-to-noise ratio using all available genetic features was low, varying thresholds were applied, selecting only genetic features for the cluster analysis occurring in specified proportions of the study cohort (supplemental Methods section 2.4). The definitive analysis was chosen by finding clear BIC/AIC signals for cluster identification. Thirty-one genetic features were used in the cluster analysis; of these, all 22 targets of somatic hypermutations were included, as well as 9 nonsomatic hypermutation features that occurred in 60 or more patients. These features are listed in supplemental Table 1 and cluster stability analysis in supplemental Methods section 2.2. aSHM targets were identified based on our prior studies in DLBCL20 and identified based on prior literature (see supplemental section 2.5).
Survival analysis used the Kaplan-Meier estimator and proportional-hazards regression, with all patients followed up for mortality until 31 December 2018. Analyses were conducted in Stata 1630 and R 4.0.331 using flexmix32 and survival33 libraries, with Benjamini-Hochberg adjustment employed for multiple comparisons.
Results
Mutation distribution in FL
Demographic and clinical characteristics of the source cohort (n = 852), the full study cohort (n = 548), and those for whom detailed clinical and treatment data were available (source cohort 830 of 852; study cohort 538 of 548) are summarized in Table 1, with the data flow outline in supplemental Figure 1.
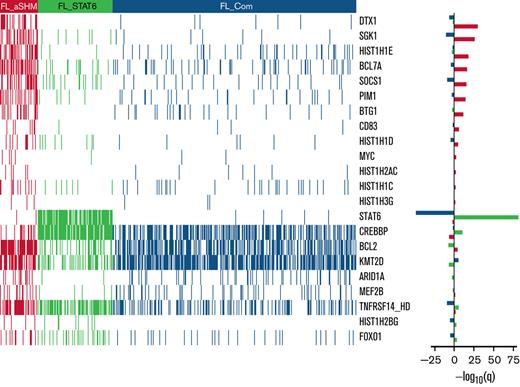
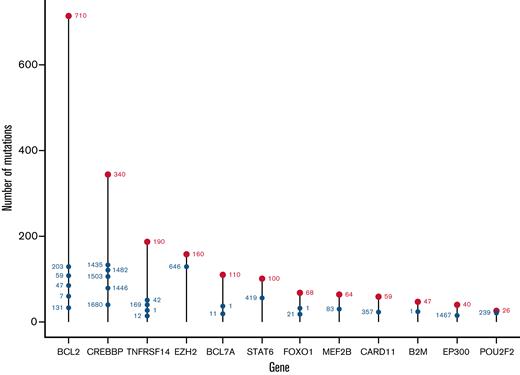
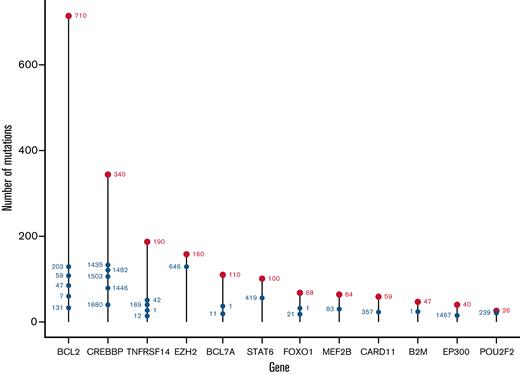
From the 293-gene panel, 181 genetic features were detected in ≥1 patient. Individual biopsies were associated with a median of 6 (interquartile range [IQR], 4-8) driver mutations, compared with a median of 7 (IQR, 5-11) in our cohort of patients with DLBCL (Figure 1A), P < 10−4.20 The most prevalent events detected were in KMT2D, CREBBP, BCL2, TNFRSF14, EZH2, STAT6, and BCL7A (Figure 1B), with specific hotspot mutations identifiable. As expected, the commonest mutational sites were EZH2 at residue 646, followed by STAT6 at residue 419, and CREBBP at 1680. Although present in lower numbers, POU2F2 (OCT2) showed the most skewed mutational hotspot; the vast majority of detected mutations occurred at residue 239 (Figure 2).
Distribution of the number of mutations. (A) Comparison of the distributions of the number of mutations per patient in FL vs DLBCL. Each bar represents the proportion of patients with FL (red) or DLBCL patients (blue) with the corresponding number of mutated genes. (B) The proportion of patients in the FL study cohort carrying the corresponding genetic feature: the top 20 most common nonaberrant somatic hypermutation genetic features. (C) The proportion of patients in the FL study cohort carrying the corresponding aberrant somatic hypermutations.
Distribution of the number of mutations. (A) Comparison of the distributions of the number of mutations per patient in FL vs DLBCL. Each bar represents the proportion of patients with FL (red) or DLBCL patients (blue) with the corresponding number of mutated genes. (B) The proportion of patients in the FL study cohort carrying the corresponding genetic feature: the top 20 most common nonaberrant somatic hypermutation genetic features. (C) The proportion of patients in the FL study cohort carrying the corresponding aberrant somatic hypermutations.
Distribution of mutation sites in the FL study cohort, by gene. The most mutated individual sites are shown for each mutated gene. The total number of mutated sites in each gene is shown by the uppermost red label; the most highly mutated sites in each gene are shown with blue labels, with their position showing the cumulative total of mutations.
Distribution of mutation sites in the FL study cohort, by gene. The most mutated individual sites are shown for each mutated gene. The total number of mutated sites in each gene is shown by the uppermost red label; the most highly mutated sites in each gene are shown with blue labels, with their position showing the cumulative total of mutations.
Using criteria previously applied to DLBCL, 22 genes were identified as potential targets of aSHM,20 many of which belonged to the most frequently mutated FL genes (Figure 1B,C). Of these, BCL2 was the commonest target. Aberrant targeting of SHM to the BCL2 gene in the context of t(14;18) is thought to reflect the recruitment of the SHM machinery by the translocated IGH regulatory elements.7 Different mechanisms of aSHM targeting are implicated in genes that are not linked by translocation to the IGH locus; these include both recruitment of AID to transcribing polymerase in strongly expressed genes and the impact of differential repair.34 Setting BCL2 aside for this reason, the most commonly mutated aSHM targets were: BCL7A, HIST1H1E, SOCS1, PIM1, HISTH1C1, BTG1, and SGK1 (Figure 1C). Considered either with or without BCL2 mutations (supplemental Figure 2A,B, respectively), FL can broadly be divided into those with or without evidence of mutation at 1 or more potential aSHM target genes. Indeed, a significant tail of cases showed evidence of mutation in multiple aSHM target genes, with a maximum of 13 mutated genes per individual patient.
FL genetic subclusters
We found that clustering FL using all available mutation data provided very weak evidence of structure. However, this could be attributable to a degree of noise in an entity showing relative homogeneity of mutational profile in comparison, for example, to DLBCL. Accordingly, we undertook a process of filtering for signal-to-noise ratio (supplemental Methods 2.4), resolving a set of 31 primary genetic features (including BCL2) that were the most informative for cluster analysis.
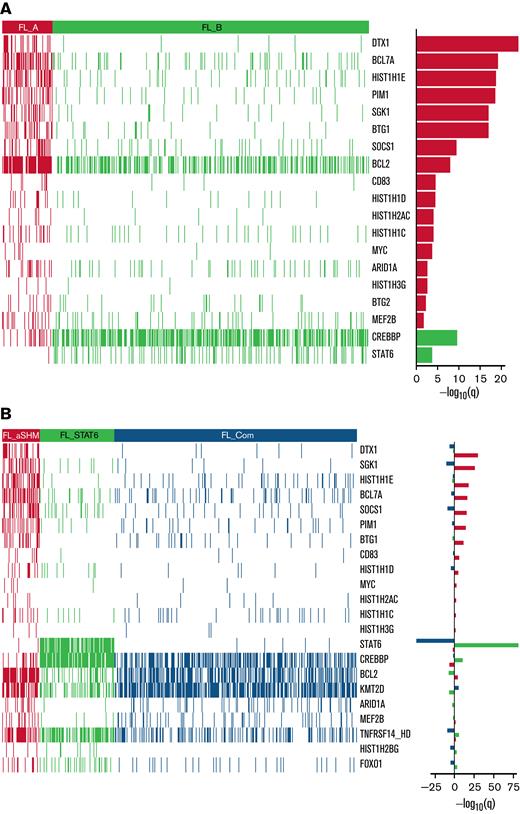
In line with our previous study,20 2 information criteria with differing stringency and sensitivity were used. The more stringent but less sensitive BIC identified 2 clusters (Figure 3A); the smaller one (FL_A; n = 75 of 548; 13.7%) was highly enriched for targets of aSHM, especially DTX1, BCL7A, HIST1H1E, PIM1, SGK1, BTG1, as well as BCL2, and the larger (FL_B; n = 473 of 548; 86.3%) for mutations of CREBBP and STAT6. The more sensitive AIC identified 3 clusters (Figure 3B). The smallest AIC cluster (n = 58 of 548; 10.6%; designated FL_aSHM) identified cases enriched for aSHM targets including DTX1, SGK1, HIST1H1E, BCL7A, SOCS1, PIM1, BTG1, as well as BCL2; the intermediate cluster (n = 115 of 548; 21.0%; designated FL_STAT6) was especially enriched for mutations of STAT6, CREBBP, and TNFRSF14; and the largest cluster (n = 375 of 548; 68.4%; designated FL_Com) was relatively enriched for mutations of KMT2D and low aSHM and absence of STAT6 mutation.
Heatmaps of mutated genes, by cluster. (A) For the 2 BIC-determined clusters, FL_A and FL_B. (B) for the AIC-determined clusters, FL_aSHM, FL_STAT6, and FL_Com. The panel on the righthand side shows the enrichment for mutations within each cluster with a logarithmic q-value scale. Only those mutations are shown that are identified as significantly enriched for the given group, as determined by a Benjamini-Hochberg adjusted q < 0.05 from a χ2 test of independence. “HD” indicates the homozygous deletion or a mutation in this gene.
Heatmaps of mutated genes, by cluster. (A) For the 2 BIC-determined clusters, FL_A and FL_B. (B) for the AIC-determined clusters, FL_aSHM, FL_STAT6, and FL_Com. The panel on the righthand side shows the enrichment for mutations within each cluster with a logarithmic q-value scale. Only those mutations are shown that are identified as significantly enriched for the given group, as determined by a Benjamini-Hochberg adjusted q < 0.05 from a χ2 test of independence. “HD” indicates the homozygous deletion or a mutation in this gene.
Supplemental Figure 3 shows the distribution of gene mutations that occurred in ≥10 patients (differentially mutated between clusters or not). Several frequently mutated genes were not significantly differentially associated with BIC or AIC clusters; notably, these include EZH2 as well as IRF8 in the context of AIC clusters.
Comparing clusters obtained using the 2 criteria, 55 of 58 cases belonging to AIC FL_aSHM also belonged to the smaller BIC FL_A cluster (Table 2). Thus, both BIC and AIC converged on a subset of FL with multiple aSHM target gene mutations. By contrast, FL_B, the larger BIC cluster, was mostly split between the FL_STAT6 and the FL_Com AIC clusters (115-355), with only 3 subjects assigned to FL_aSHM AIC. Segregation of an additional AIC cluster was reflected in the separation of a subgroup of cases principally characterized by STAT6 mutation (FL_STAT6). Of note, in an independent data set,35 clustering with AIC rediscovered the separation of FL cases with high aSHM burden from those without (supplemental Methods section 2.3). In this external dataset, cases with STAT6 mutation were not present at sufficient frequency to allow their separation. This may be due to the inclusion criteria and skewing of STAT6 mutation in relation to clinical stage and associated treatment choice.
Translocation of BCL2 and, to a lesser extent, BCL6, are key pathogenetic events in FL. Our approach focused specifically on the question of clustering based on gene mutation rather than structural genomic features. Among cases with translocation data accrued during routine diagnosis, we found no evidence of bias between the mutation-defined subgroups for the absence/presence of a detected BCL2 translocation (BCL2 translocation positive/tested: FL_aSHM 21/25; FL_STAT6 25/32; FL_Com 101/114). We note that this does not exclude the potential for selective enrichment of STAT6 mutation among BCL2 translocation-negative FL, as has been previously reported, but argues that STAT6 mutation may occur as a feature of both BCL2 translocation-positive and -negative FL cases.36-39
Mutational profiles in FL subgroups
BIC clustering only on aSHM targets (excluding BCL2) confirmed the segregation based on these features alone, supporting the conclusion that a significant subset of FL is characterized by a mutational pattern consistent with higher aSHM activity. Seeking further evidence of differential mutational mechanisms between FL subsets, we carried out a mutational signature analysis. Although mutational signatures are conventionally analyzed at the whole genome sequencing level, we reasoned that mutational mechanisms acting at relevant oncogenes/tumor suppressors are of particular interest, and our data, while biased in terms of genomic coverage, allowed meaningful comparison between cases and sets of genes in the panel. Therefore, all single base substitutions across the tiled regions in the target panel were considered across the set of FL cases and analyzed using the Signal platform,29 providing a case-by-case representation of substitutions in their triplet context.
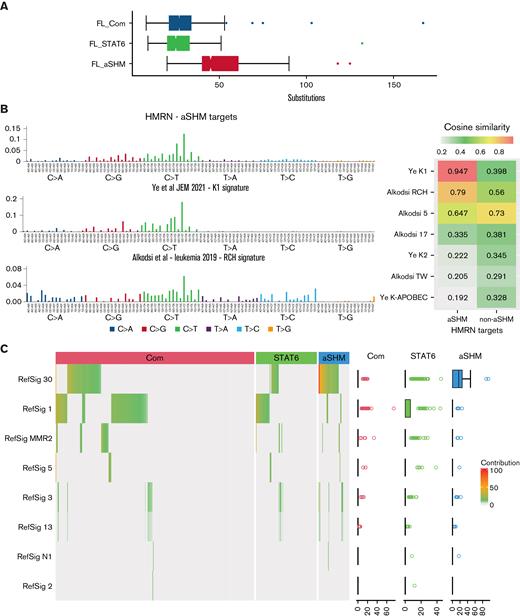
A wide range of substitutions per case (8-167) was observed, and the absolute substitution burden was skewed regardless of mutation signature between the FL subgroups (Figure 4A). Aggregating all the substitutions observed across groups of genes increased the depth of data across tiled regions and demonstrated that the pattern of substitution differed between the genes defined as targets of aSHM relative to the substitution pattern across the remainder of the genes used in our clustering analysis, or the extended group of all lymphoma genes not defined as aSHM targets (supplemental Figure 5). The pattern of substitutions at aSHM targets was similar to that previously described as aSHM targets genome-wide in the K1 signature by Ye and colleagues40 and also related to the RCH signature41 with a preference for G(C>T)T and G(C>T)C conforming to the RCY context (R = A/G, Y = C/T) that characterizes the core of SHM hotspots (Figure 4B).
Distribution of substitutions and predicted mutation signatures. (A) Comparison of the distributions of the number of substitutions per patient in the cohort divided according to AIC cluster as indicated in the figure. (B) Left: pattern of substitutions in triplet context: observed across the cohort for genes identified as aSHM targets (upper panel), in the K1 signature described by Ye et al40 (middle panel), and in the RCH signature described by Alkodsi et al41 (lower panel). The substitution type is indicated in the color-coded squares below the graph, with individual triplet contexts plotted by relative substitution frequency with matching color-coded bars. Right: cosine similarity measures. (C) Predicted mutational Reference Signature (RefSig) contribution to the substitution pattern observed in each sample using the Signal analysis package is displayed as a hierarchically clustered heatmap, divided by BIC FL clustering (color-coded bars above the heatmap). The percentage contribution of the predicted RefSig is indicated in the white (0) to red (100) color scale, as shown in the figure. Predicted contributing signatures are shown to the left. The distribution of predicted contributing signatures is shown to the right as a scatterplot. Note mutation signatures were derived from patterns observed in the lymphoma driver panel; cases with low mutation burden fall below the threshold for signature prediction.
Distribution of substitutions and predicted mutation signatures. (A) Comparison of the distributions of the number of substitutions per patient in the cohort divided according to AIC cluster as indicated in the figure. (B) Left: pattern of substitutions in triplet context: observed across the cohort for genes identified as aSHM targets (upper panel), in the K1 signature described by Ye et al40 (middle panel), and in the RCH signature described by Alkodsi et al41 (lower panel). The substitution type is indicated in the color-coded squares below the graph, with individual triplet contexts plotted by relative substitution frequency with matching color-coded bars. Right: cosine similarity measures. (C) Predicted mutational Reference Signature (RefSig) contribution to the substitution pattern observed in each sample using the Signal analysis package is displayed as a hierarchically clustered heatmap, divided by BIC FL clustering (color-coded bars above the heatmap). The percentage contribution of the predicted RefSig is indicated in the white (0) to red (100) color scale, as shown in the figure. Predicted contributing signatures are shown to the left. The distribution of predicted contributing signatures is shown to the right as a scatterplot. Note mutation signatures were derived from patterns observed in the lymphoma driver panel; cases with low mutation burden fall below the threshold for signature prediction.
The aSHM K1 signature demonstrated a cosine similarity of 0.947 relative to the observed substitution pattern at aSHM target genes in our FL cases, contrasting with a cosine similarity of 0.398 for the substitution pattern at non-aSHM targets (Figure 4B). Among predicted contributions of reference pan-cancer mutation signatures provided by Signal, the aSHM targets differed from other genes in the panel, with marked skewing to the RefSig30 mutation signature (predicted contribution: 69% at aSHM; 17% at non-aSHM clustering genes; 25% at all non-aSHM panel genes) (supplemental Figure 5). While the number of mutations observed in the targeted panel was limiting for many cases, an estimated contribution of reference pan-cancer mutation signatures could be predicted for a substantial subset of individual cases. We, therefore, used these predictions to assess whether the observed skewing between FL clusters was maintained at the level of individual cases. RefSig30 was the dominant mutational signature assigned to cases in the FL_aSHM subgroup, while FL_Com cases showed a mixed pattern predominantly reflecting RefSig_1 as well as the admixture of RefSig30 in some cases (Figure 4C). By contrast, FL_STAT6 was enriched for cases with a predicted contribution of RefSig1 but not RefSig30. We acknowledge the limitation inherent in assessing mutational signatures using targeted sequencing panels relative to whole genome or exome studies. However, we reason that individual cases are treated equally in this analysis, allowing conclusions to be drawn from comparative analysis regarding the potential mutational processes and detected mutation frequency at a relevant set of genomic regions. This demonstrates that the subgroup of FL_aSHM cases is distinguished from both FL_STAT6 and FL_Com by a higher absolute burden of substitutions, with the overall substitutions pattern at FL_aSHM target genes showing a dominant pattern with near identity (cosine similarity 0.947) to the “AID fingerprint” pattern of aSHM at kataegis regions described previously by Ye and colleagues.40 This pattern is also closely related to the cAID pattern identified by Chapuy and colleagues in DLBCL,18 and the SHM-like mutational pattern at IG-VDJ regions described by Hubschmann and colleagues.42 In contrast, FL_STAT6 cases have a lower overall substitution burden across our lymphoma oncogene panel, showing little evidence of aSHM-related mutation patterns in these key oncogenic regions. Together these data support the conclusion that the FL_aSHM subgroup identifies a distinct etiological subgroup at the level of mutation signature.
STAT6 mutant FL
Recurrent STAT6 mutations have been previously characterized in FL.12,14 Here, the separation of FL clusters characterized by STAT6 mutation using the AIC and the difference in mutation pattern observed in such cases provides strong evidence for this as a distinct entity. Of the 100 patients with STAT6 mutation, 99 had a single mutation, with 1 patient having 2. Fifty-six of 101 mutations fell at the 419D hotspot, with 24 of 101 being D/G mutations, followed by 13 of 101 D/N and 9 of 101 D/H (supplemental Figure 6). The second most frequent (14 of 101) was at position 377 E/K, with 7 of 101 at 372 E/K. These patterns are consistent with those previously described, demonstrating the enrichment for mutations affecting residues associated with the STAT6 DNA-binding domain.14 This is a feature of both FL and other B-cell lymphomas with STAT6 mutations and is associated with STAT6 activation.14,43 Hence, the FL_STAT6 cluster defined by STAT6 mutation in our data is consistent with a subgroup characterized by deregulated activation of this pathway and low levels of aSHM.
It has been suggested that STAT6 and CREBBP mutations may represent a cooperative mutational pairing in FL,14 a proposal that our AIC clustering supports. Examining patterns of cooccurrence of mutations in STAT6 and the other 5 commonest targets (KMT2D, CREBBP, BCL2, TNRFS14, and EZH2_646) independently of clustering confirmed that STAT6 mutation occurs more frequently than expected with CREBBP mutation. This was evident both for the STAT6/CREBBP pairing alone (adjusted P = .018), the combination of STAT6/CREBBP with TNFRSF14 (adjusted P = .0043), and to a lesser extent TNFRSF14 and EZH2_646 (adjusted P = .031). Furthermore, STAT6 and BCL2 comutation (adjusted P = .022) and STAT6, BCL2, and KMT2D comutation (adjusted P = .013) were significantly less common than expected if mutations were independent. In the absence of STAT6 mutation, the fourfold combination of EZH2_646, BCL2, KMT2D, and TNFRSF14 mutation occurred more commonly (adjusted P = .031), as did the fivefold combination including CREBBP (adjusted P = .027). However, these combinations were less frequent and enrichments less significant than the 2 commonest STAT6 patterns. The combinations of STAT6/CREBBP and STAT6/CREBBP/TNFRSF14 provide the strongest evidence for significant coenrichment among dominant mutational targets, further supporting the validity of the STAT6-, CREBBP-, and TNFRSF14-enriched FL_STAT6 clusters.
Demographics and clinical features of AIC subclusters
No statistically significant differences in sex or FLIPI were evident (Table 3). However, patients in FL_aSHM were, on average, 4.8 years older than those in the other 2 clusters (P = .007). Stage distributions also differ: FL_aSHM has a comparative excess of stage III, FL_STAT6 of stage I and II, and FL_Com of stage IV. As might be expected given this, the underpinning frequency of documented bone marrow involvement at diagnosis in FL_Com was also higher: 204 of 375 (54%) and 63 of 173 (36%), respectively (P < .001). This was not attributable to differences in bone marrow investigation, which were, by contrast, skewed in favor of FL_aSHM (91.23%) compared with FL_Com (85.29%) (supplemental Methods section 2.8).
Overall survival (OS) was lower for FL_aSHM relative to FL_Com (Figure 5A) (P = .03), but this did not persist after adjustment for age at diagnosis, and no differences were evident for BIC clusters (Figure 5B). Although no survival differences were detected for patients initially managed by “Watch and Wait” (Figure 5C,D), AIC cluster differences were detected in those initially managed in other ways (listed in Table 1) (P = .04), with weak evidence for BIC clusters (P = .08) (Figure 5E,F). As before, neither of these differences persisted following age adjustment.20
OS of all patients with follow-up data, by cluster. (A) OS of patients by BIC cluster. (B) OS of patients by AIC cluster. (C) OS of patients initially put on “Watch-and-Wait” therapy by BIC cluster. (D) OS of patients initially put on “Watch-and-Wait” therapy by AIC cluster. (E) OS of patients initially treated with chemotherapy by BIC cluster. (F) OS of patients initially treated with chemotherapy by AIC cluster.
OS of all patients with follow-up data, by cluster. (A) OS of patients by BIC cluster. (B) OS of patients by AIC cluster. (C) OS of patients initially put on “Watch-and-Wait” therapy by BIC cluster. (D) OS of patients initially put on “Watch-and-Wait” therapy by AIC cluster. (E) OS of patients initially treated with chemotherapy by BIC cluster. (F) OS of patients initially treated with chemotherapy by AIC cluster.
BIC clusters were predictive of POD24 status44 (odds ratio [OR], 2.78; 95% confidence interval [CI], 1.09-7.14) in patients initially treated with R-CVP, an effect that persisted after age adjustment (OR, 3.03; 95% CI, 1.15-7.69) (supplemental methods section 2.6). Adverse POD24 was associated with the FL_A subgroup (n = 34), which includes 26 of 27 cases in the AIC FL_aSHM subgroup as well as an additional 8 cases that AIC assigns to FL_Com, indicating that the observed survival difference is not robust to clustering method.
Univariate OS modeling identified 5 genes with significant adverse hazard ratios (HRs): HIST1H2AC (HR, 3.10; 95% CI, 1.64-5.86) and BCL2 (HR, 1.45; 95% CI, 1.10-1.93), as well as ARID1A (HR, 1.84; 95% CI, 1.27-2.67), CDKN2A (HR, 2.26; 95% CI, 1.31-3.89), and TP53 (HR, 1.75; 95% CI, 1.06-2.88). After FLIPI adjustment, only the ARID1A effect persisted (HR, 1.73; 95% CI, 1.15-2.60). Evaluation of m7-FLIPI showed no enhancement over stratifying survival by FLIPI and ECOG Performance Status alone (supplemental Methods section 2.7). Hence, separation of FL according to mutational status despite being linked to apparent underlying mechanistic differences provides limited prognostic information in conventionally treated patients.
FL heterogeneity and transformation
Ninety-six of 548 FL cases transformed to DLBCL during follow-up (Table 1). Over a quarter of these (26 of 96) had samples taken at both timepoints, but no evidence of transformation imbalance was found (FL_aSHM = 13.8%; FL_STAT6 = 17.4%; FL_Com = 18.1% transformed; P = .72), and cluster was not predictive of time-to-transformation (P = .9). However, univariate modeling of time-to-transformation to DLBCL identified 2 genes with adverse HRs: TP53 (HR, 3.13; 95% CI, 1.71-5.76) and CDKN2A (HR, 2.50; 95% CI, 1.16-5.41) (CDKN2A only had 7 transformations to DLBCL in the mutated arm). BTK (HR, 2.07; 95% CI, 0.96-4.48; P = .064) and GNA13 (HR, 1.76; 95% CI, 0.94-3.29; P = .080) showed weak evidence of an association. One gene, IRF8 (HR, 0.50; 95% CI, 0.23-1.07; P = .074), showed weak evidence of an association with a favorable risk. Time-to-transformation was not associated with the burden of aSHM, but there was weak evidence of association with non-aSHM (P = .052).
Genetic abnormality frequencies were assessed for the 26 subjects with paired FL and DLBCL samples (Figure 6; supplement Figure 7): 17 of 26 had ≥2 unique to the DLBCL sample, with a maximum of 8 additional genetic abnormalities detected. We considered that there was evidence of branching evolution where mutations unique to both FL and DLBCL were identified, along with a set of shared mutations in both samples, and in only 9 of 26 was this evidence lacking. Analysis of variant allele fraction (vAF) (Figure 7; supplemental Figure 8) for shared and disparate mutations between samples further supports the maintenance of high vAFs for common clonal mutations alongside the emergence of new mutations that were not identified at first diagnosis, consistent with prior publications.12,15,23,45,46 Examining mutations across individual FL and DLBCL pairs (Figure 7) demonstrated common retention of core oncogenic FL events, including mutation of BCL2, KMT2D, CREBBP, and STAT6 on transformation.
Mutations in patients who transformed from FL to DLBCL. Distribution of mutations by individual patient stratified by mutated at FL diagnosis only, mutated at DLBCL only, and mutated at both FL and DLBCL diagnosis.
Mutations in patients who transformed from FL to DLBCL. Distribution of mutations by individual patient stratified by mutated at FL diagnosis only, mutated at DLBCL only, and mutated at both FL and DLBCL diagnosis.
Variant Allele Frequency Trajectories. (A-D) Show variant allele frequency trajectories (for samples taken at diagnosis with FL and with DLBCL) for a selection of individual patients (numbers 6, 7, 25, and 26, respectively, in [A-D]). Gene/sites are annotated. Patient 6 shows a complex set of gains with selection for HLA-B mutations; patient 7 shows a complex set of gains; patient 25 shows substantial shared mutation burdens, with a couple of gains from zero and one loss and Includes EZH2 as a shared mutation; patient 26 shows a core set of 5 mutations and major gain in CREBBP.
Variant Allele Frequency Trajectories. (A-D) Show variant allele frequency trajectories (for samples taken at diagnosis with FL and with DLBCL) for a selection of individual patients (numbers 6, 7, 25, and 26, respectively, in [A-D]). Gene/sites are annotated. Patient 6 shows a complex set of gains with selection for HLA-B mutations; patient 7 shows a complex set of gains; patient 25 shows substantial shared mutation burdens, with a couple of gains from zero and one loss and Includes EZH2 as a shared mutation; patient 26 shows a core set of 5 mutations and major gain in CREBBP.
Discussion
Our analysis of 548 patients with FL from a United Kingdom population-based cohort supports the existence of significant nonrandom patterns of cooccurring mutations. These findings are consistent with previous studies examining the most common targets of mutation in FL, underlining the importance of CREBBP, KMT2D, and TNFRSF14. Similarly, hotspot mutation of EZH2-646 was the single most common mutated residue in FL, followed quite closely by hotspot mutation in STAT6-419, highlighting the previously suggested importance of this site.14
Our clustering approach demonstrated that FL could be resolved into mutational subgroups with specific mechanistic differences. FL_aSHM links to high levels of aSHM; FL_STAT6 identifies STAT6 mutated cases as a distinct cluster, which was significantly associated with CREBBP alone, and in combination with TNFRSF14. FL_Com was enriched for KMT2D mutation but was principally defined by the absence of the enriched mutations in FL_STAT6 and low mutation frequencies of aSHM targets. Because we focused on a preselected target panel that was matched to our previous DLBCL study,20 we cannot exclude the possibility that FL_Com may be further subdivided by mutation at targets that are either missing from our panel or are in noncoding regions. In particular, RRAGC, ATP6V1B2, VMA21, as well as CTSS and EEF1A1 have emerged as important mutations in FL, with frequency estimates varying from <10% to <20%,47,48 and in the case of EEF1A1, for example, being enriched in the duodenal subtype of FL which is underrepresented in our study.15,46 We note that ATP6AP1, another gene linked to RRAGC, was included in the extended regions of our panel but was only identified as mutated in a single FL case. STAT6 mutation has been found to be enriched in selected series of FL with diffuse growth pattern, CD23 expression, and/or absence of t(14;18).36-39 Our analysis identifies the FL_STAT6 cluster independent of such criteria but sharing associated molecular features, providing further evidence for this as a distinct molecular subtype of FL. In these prior studies of FL enrichment of STAT6 with CREBBP and TNFRSF14 mutation has been a consistent finding which is shared with our analysis. None of these prior studies appear to have found coenrichment of mutations in RRAGC, ATP6V1B2, VMA21, CTSS, or EEF1A1, arguing that the mutation status of this group of genes though interesting is unlikely to impact FL_STAT6 membership. In terms of the FL_aSHM subgroup, we have shown that this is identifiable in an additional external independent data set, which includes mutational information on EEF1A1 and ATP6V1B2, neither of which showed significant effects on clustering in this data. It is unlikely that the relatively rare mutation of genes outside our targeted panel would impact the dominant effect of the aSHM mutation pattern, but these may have importance in the context of subdivision within the FL_Com group.
That high aSHM load is the most robust feature subdividing a significant subset of FL is notable, implying that either targeting or resolution of the aSHM process is deregulated in these cases.34 Analysis of mutational signatures provides independent evidence of the mutagenic process, separate from the assumption of aSHM targeting based on prior evidence. The mutational pattern observed both at the specific subsets of presumptive aSHM targets across our FL case series and within the individual FL_aSHM cases across all substitutions detected is closely related to that identified for the aSHM process in previous genome-wide studies.42 The observed substitution preference for G(C>T)T and G(C>T)C conforms to the RCY context (R = A/G, Y = C/T) that characterizes the core of SHM hotspots. An interesting feature is that this mutation pattern and mutation of target genes that contribute to the FL_aSHM subgroup are more frequently observed in DLBCL.20,40 However, we were unable to show a strong association between FL_aSHM and risk of transformation. While aSHM presents a common etiological mechanism, it is likely that the consequent mutations in individual cases are of significance in determining clinical behavior; indeed, in the context of DLBCL, mutations in SOCS1 and SGK1 contribute to the identification of a specific subset, and separate from the alternate aSHM target BTG1, while all of these genes contribute to the identification of the FL_aSHM subgroup. It may be of particular interest to note mutations in SGK1 that lead to the expression of stable active truncated isoforms.49 Additionally, several histone H1 encoding genes, including HIST1H1E, are among the most frequent targets associated with FL_aSHM, supporting a common mechanism of downstream epigenetic deregulation. Recent work has illustrated the profound effect of histone H1 mutation on chromatin structure and oncogenesis.50 Mutation of HIST1H2AC may act in a related fashion since alterations in histone H2A usage have also been implicated as common oncogenic events in cancer.51 Further evaluation will be needed to assess the potential association between aSHM and histone-mediated epigenetic perturbation, but this may represent a frequently shared characteristic of the FL_aSHM subset.
The association between mutations targeting DNA-damage responses (TP53 and CDKN2A) and risk of transformation makes intuitive sense since these pathways are well characterized in aggressive lymphoma and cancer progression more generally. Furthermore, copy number alterations affecting TP53 and CDKN2A loci have been linked to adverse outcomes in FL.52 The weak associations of GNA13 and BTK mutations with shorter time-to-transformation, and IRF8 with longer time-to-transformation, are suggestive of an association with potential pathways linked to GC biology.53-55 Of these, loss of GNA13 has been identified as strongly cooperative with BCL2 and BCL6 oncogene expression in the transformation of human in vitro lymphoma models,56 and has been implicated as a gene with an enhanced mutation in transformed FL.15 However, even in our large cohort, the low rate of progression limited statistical power, and cases lacking these features also transformed. This could also explain the lack of evidence supporting an association between mutational clusters and transformation.
An important difference between our analyses of FL and DLBCL is that while recurrent patterns of mutation, in keeping with differences in disease biology, can be identified in both, these show limited prognostic significance in our FL data. In part, this reflects a diversity of management strategies and the intrinsically indolent nature of the FL disease process. However, overall our analyses suggest that mutational stratification has limited prognostic value in FL and that in terms of clinical response, these mutational combinations generate phenocopies in reaction to conventional treatment strategies. The underlying mechanistic difference may, however, offer opportunities for the development of future treatment strategies related to molecular subtypes.
We acknowledged that the sequencing was based on tumor-only FFPE samples. Even with very stringent filtering, some germline mutations and sequencing artifacts cannot be excluded from such mutational analyses. However, we note that these are not likely to significantly impact the identification of driver mutation calls or the identification of mutational signatures linked to specific patterns of base substitution.
In conclusion, our population-based analysis of mutation patterns in FL provides evidence in support of distinct mechanistic entities, with aSHM burden and STAT6 mutation status providing key determinants of this molecular heterogeneity. In contrast to DLBCL, we did not find evidence that tumor genetics is a major determinant of patient outcome in FL. Instead, we speculate that tumor-extrinsic factors related to the microenvironment or host immune system may have a greater influence on patient survival.
Acknowledgments
Blood Cancer UK (grant number 15037), Cancer Research UK (grant number A29685), and the National Institute for Health Research (grant number RP-PG-0613-20002) funded the majority of this study. Genetic sequencing was funded by 14M Genomics, a start-up company that ceased trading in February 2016. The National Institutes of Health Research, Blood Cancer UK, and Cancer Research UK had no role in study design, data collection, data analysis, data interpretation, or report writing. 14M Genomics was involved in gene panel design but took no further part.
Authorship
Contribution: S.L.B., P.A.B., D.P., A.S., E.R., P.J.C., R.M.T., R.P., C.B., and S.C. conceptualized and designed the study; S.L.B., P.A.B., D.P., A.S., E.R., S.L.C., P.J.C., R.P., C.B., S.C., P.G., S.J.L.v.H., and N.W. provided the provision of study material or patients; S.C., D.P., R.M.T., S.L.B., P.A.B., A.S., E.R., S.L.C., P.J.C., C.R., R.P., C.B., D.J.H., and S.E.L. analyzed and interpreted the data; R.M.T., S.C., A.S., and E.R. wrote the manuscript; and all authors collected and assembled data and approved the final draft of the manuscript.
Conflict-of-interest disclosure: P.A.B. is the director at Tessellex Ltd and Gabriel Precision Oncology Ltd. D.J.H. receives research funding from Gilead International and provides consultancy for Karus. The remaining authors declare no competing financial interests.
The current affiliations for C.R. are the Department of Bioengineering and the Department of Computer Science, Stanford University, Palo Alto, CA. P.A.B. is currently affiliated with Hull York Medical School, Heslington, York.
Correspondence: Simon Crouch, Epidemiology and Cancer Statistics Group, Department of Health Sciences, University of York, York, YO10 5DD, United Kingdom; e-mail: simon.crouch@york.ac.uk.
References
Author notes
The mutation data features are in supplemental Table 6. The sequencing data can be found in the European Genome-Phenome Archive (https://ega-archive.org/, acquisition number: EGAS00001005238). The R-code is available at https://github.com/ecsg-uoy/FLGenomicSubtyping. Please contact the corresponding author for other data sharing at simon.crouch@york.ac.uk.
S.C., D.P., S.L.B., and P.A.B. are joint first authors.
C.B., A.S., and R.M.T. are joint last authors.




![Variant Allele Frequency Trajectories. (A-D) Show variant allele frequency trajectories (for samples taken at diagnosis with FL and with DLBCL) for a selection of individual patients (numbers 6, 7, 25, and 26, respectively, in [A-D]). Gene/sites are annotated. Patient 6 shows a complex set of gains with selection for HLA-B mutations; patient 7 shows a complex set of gains; patient 25 shows substantial shared mutation burdens, with a couple of gains from zero and one loss and Includes EZH2 as a shared mutation; patient 26 shows a core set of 5 mutations and major gain in CREBBP.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/21/10.1182_bloodadvances.2021005284/7/m_blooda_adv-2021-005284-gr7.jpeg?Expires=1765126519&Signature=ou4hE86zrMiRdKHkfMIpwiUcVPQne-PKJnH~9OGDnXUjTd3WQ0rTsZzNwlZcRleNHlzCbncb~OXP4qepLjmAmUeuRy~6foUMzpjPq0xb7zqP7MUm42BDfhk9V2cqPhQlW-nm3~usseZnVBXRqq~lXST5FSRi3JGQehVupkQWHaswGeDOq1Zof5KjJTtDZs2bf259ASk-Iuubj-MOre7VEesdGHMPLMWQR7dWJ2eoa7n1BFeH1bwgb7IzLO088Qr6w0mq1tSIEdUDs6R1e8gGFBHx9dDn~-7MLY1yEz2Uepi6zz9kxeoqBABjwyBuu0u2jkoTSfoP5wOywZfrVSFmtQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)







![Variant Allele Frequency Trajectories. (A-D) Show variant allele frequency trajectories (for samples taken at diagnosis with FL and with DLBCL) for a selection of individual patients (numbers 6, 7, 25, and 26, respectively, in [A-D]). Gene/sites are annotated. Patient 6 shows a complex set of gains with selection for HLA-B mutations; patient 7 shows a complex set of gains; patient 25 shows substantial shared mutation burdens, with a couple of gains from zero and one loss and Includes EZH2 as a shared mutation; patient 26 shows a core set of 5 mutations and major gain in CREBBP.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/21/10.1182_bloodadvances.2021005284/7/m_blooda_adv-2021-005284-gr7.jpeg?Expires=1765126520&Signature=ALFzSqLzvNsO9ABfEJi0NBeztbcqmNai6CLvBTbC9e-6IY7Ask4TZHWTI2oDIV9VKb8Iw35xVOrQnXePh5l-Z44LSeOraa5y6h5GxV6AyoafvBaQD5alf6NvkwH71H6h76GpXy2Mvz8fDKxqrDXXJpkGVmSc7JpJivaIi~IbKkDot8AZoe-P6cthS0to~loBfHTRJNpXQWlFzr7yZkbZg8N8IZSQdiQkacIHm2NQuuKq45BI3KMqqDmtLTYvozxl25KAdwTpjfstZDqdJmRGtJ3JGkrjddsSxhBAEZgYQmZKFdKlhclYP6w-EqiaS4G6LiZct-pkhIJuAeLXx00TJw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)