This glossary is designed to help the reader with the terminology of molecular biology. Each year, the glossary will be expanded to include new terms introduced in the Education Program. The basic terminology of molecular biology is also included. The glossary is divided into several general sections. A cross-reference guide is included to direct readers to the terms they are interested in. The hope is that this addition to the Education Program will further the understanding of those who are less familiar with the discipline of molecular biology.TBL1
Cross-Reference Guide
| Term . | Section . |
|---|---|
| Actinomycin D pulse experiments | V |
| Adeno-associated viral vectors | VIII |
| Adenoviral vectors | VIII |
| ALK | X |
| Allele-specific hybridization | XI |
| Allele-specific PCR | IV |
| AML-1 | X |
| Amphotropic virus | VIII |
| Anaplastic lymphoma kinase | X |
| Antisense oligonucleotides | VIII |
| Basic helix-loop-helix proteins | V |
| Bcl-1 | X |
| Bcl-2 | X |
| Bcl-3 | X |
| Bcl-6 | X |
| β galactosidase | V |
| Branched chain DNA signal amplification assay | II |
| c-abl | X |
| c-fos | X |
| c-jun | X |
| c-myb | X |
| c-myc | X |
| c-ras | X |
| c-rel | X |
| Calcium phosphate | VI |
| CAN | X |
| CAT | V |
| cDNA | II |
| cDNA blunting | IX |
| cDNA library preparation | IX |
| cdk | V |
| cdkI | V |
| CFBβ | IX |
| Chimeraplasty | VIII |
| Chitosan-DNA | VIII |
| Chloramphenicol transferase | V |
| Chromatography, gel filtration | IV |
| Chromatography, ion exchange | IV |
| Chromatography, hydrophobic | IV |
| Chromatography, affinity | IV |
| Chromatography, high performance liquid (HPLC) | IV |
| Cis-acting factors | V |
| Codon | II |
| Color complementation assay | XI |
| Comparative gene hybridization | IV |
| Competitive oligonucleotide hybridization | XI |
| Concatamerization | VI |
| Cyclin-dependent kinase | V |
| Contig | VII |
| Cosmid | II |
| CpG nucleotide | II |
| Cyclins | V |
| DEAE dextran | VI |
| DEK | X |
| Dideoxynucleotide (ddN) chain termination sequencing | IV |
| Directional cloning | IX |
| DNA (deoxyribonucleic acid) | II |
| DNA methylases | III |
| DNA polymerase | III |
| DNAse footprinting | IV |
| DNAse hypersensitivity site mapping | IV |
| Ecotropic vectors | VIII |
| Ecotropic virus | VIII |
| Electroporation | VI |
| Endonuclease | III |
| Enhancer | V |
| Episomal | VIII |
| ETO | X |
| Evi-1 | X |
| Exons | V |
| Exonuclease | III |
| Farnesyl protein transferase | III |
| Fas | X |
| First strand synthesis | IX |
| FISH (fluorescence in situ hybridization) | IV |
| FTPase | III |
| Gene knock-in experiments | VII |
| Gene knock-out experiments | VII |
| Helix-turn-helix | V |
| Homologous recombination | VII |
| Hox II | IX |
| HPLC | IV |
| In situ hybridization | IV |
| Initiation codon | V |
| Initiation complex | V |
| Interferon regulatory factor | X |
| Introns | V |
| IRF-1 | X |
| IRF-2 | X |
| Isoschizomer | III |
| Kinases | III |
| Klenow fragment | III |
| KOZAK sequence | V |
| LCR | V |
| Leucine zipper proteins | V |
| Library screening | IX |
| Ligases | III |
| Linkering | IX |
| Liposomes | VI |
| Locus control region | V |
| Long terminal repeat | VIII |
| Luciferase | V |
| Mammalian protein kinases | III |
| Master switch genes | V |
| Max | X |
| Maxam-Gilbert sequencing | IV |
| Minimal residual disease | IV |
| Missense mutation | V |
| MLL | X |
| Mobility shift (or band shift) assays | IV |
| mRNA | II |
| Mutagenesis, site-specific | IV |
| Nested PCR | IV |
| NF-1 | X |
| Nick-translation | IV |
| Nonsense mutation | V |
| Nonviral transduction methods | VIII |
| Northern blotting | IV |
| Nucleases | III |
| Nucleosomes | V |
| ORF (open reading frame) | II |
| p53 | X |
| PCR (polymerase chain reaction) | IV |
| Phage | II |
| Plasmids | II |
| Polyadenylation | V |
| Polylysine-ligand DNA | IX |
| Polymerases | III |
| Positional variegation | VIII |
| Post transcriptional regulation | V |
| Protein translation | V |
| Pseudotype retroviral vectors | IV |
| Pseudotyped viruses | VIII |
| Random priming | IV |
| RAR | X |
| Rb | X |
| RDA (representational difference analysis) | IX |
| Real-time PCR | IV |
| Reporter genes | V |
| Restriction endonuclease | III |
| Restriction fragment length polymorphism | XI |
| Retinoic acid receptor | X |
| Retroviral vectors | VIII |
| Reverse allele-specific hybridization | XI |
| Reverse genetics | IX |
| Reverse PCR | IV |
| Reverse transcriptase | III |
| RFLP | XI |
| Ribonuclease | III |
| Riboprobes | IV |
| Ribozyme | III |
| RNA (Ribonucleic acid) | II |
| RNA polymerase II | III |
| RNA polymerase III | III |
| RNAse protection assay | IV |
| S1 nuclease analysis | IV |
| SCL | X |
| Second strand synthesis | IX |
| Silencer | V |
| Southern blotting | IV |
| Southwestern blotting | IV |
| Splicing | V |
| Subtractive library | IX |
| Tal-1 | X |
| TATA | V |
| Tel | X |
| Telomere | II |
| Telomerase | III |
| Terminal deoxynucleotidyl | III |
| Thermostabile polymerases | III |
| Topoisomerase | III |
| Trans-acting factors | V |
| Transcription | V |
| Transcription factors | V |
| Transcriptional regulation | V |
| Transduction | VI |
| Transfection | VI |
| Transgenic animals | VIII |
| Transposon | VII |
| tRNA | II |
| Viral-derived kinases | III |
| Viral-derived transduction vectors | VIII |
| Western blotting | IV |
| X-linked methylation patterns | XI |
| YAC | VII |
| Yeast artificial chromosome | VII |
| Zinc finger domain proteins | V |
| Term . | Section . |
|---|---|
| Actinomycin D pulse experiments | V |
| Adeno-associated viral vectors | VIII |
| Adenoviral vectors | VIII |
| ALK | X |
| Allele-specific hybridization | XI |
| Allele-specific PCR | IV |
| AML-1 | X |
| Amphotropic virus | VIII |
| Anaplastic lymphoma kinase | X |
| Antisense oligonucleotides | VIII |
| Basic helix-loop-helix proteins | V |
| Bcl-1 | X |
| Bcl-2 | X |
| Bcl-3 | X |
| Bcl-6 | X |
| β galactosidase | V |
| Branched chain DNA signal amplification assay | II |
| c-abl | X |
| c-fos | X |
| c-jun | X |
| c-myb | X |
| c-myc | X |
| c-ras | X |
| c-rel | X |
| Calcium phosphate | VI |
| CAN | X |
| CAT | V |
| cDNA | II |
| cDNA blunting | IX |
| cDNA library preparation | IX |
| cdk | V |
| cdkI | V |
| CFBβ | IX |
| Chimeraplasty | VIII |
| Chitosan-DNA | VIII |
| Chloramphenicol transferase | V |
| Chromatography, gel filtration | IV |
| Chromatography, ion exchange | IV |
| Chromatography, hydrophobic | IV |
| Chromatography, affinity | IV |
| Chromatography, high performance liquid (HPLC) | IV |
| Cis-acting factors | V |
| Codon | II |
| Color complementation assay | XI |
| Comparative gene hybridization | IV |
| Competitive oligonucleotide hybridization | XI |
| Concatamerization | VI |
| Cyclin-dependent kinase | V |
| Contig | VII |
| Cosmid | II |
| CpG nucleotide | II |
| Cyclins | V |
| DEAE dextran | VI |
| DEK | X |
| Dideoxynucleotide (ddN) chain termination sequencing | IV |
| Directional cloning | IX |
| DNA (deoxyribonucleic acid) | II |
| DNA methylases | III |
| DNA polymerase | III |
| DNAse footprinting | IV |
| DNAse hypersensitivity site mapping | IV |
| Ecotropic vectors | VIII |
| Ecotropic virus | VIII |
| Electroporation | VI |
| Endonuclease | III |
| Enhancer | V |
| Episomal | VIII |
| ETO | X |
| Evi-1 | X |
| Exons | V |
| Exonuclease | III |
| Farnesyl protein transferase | III |
| Fas | X |
| First strand synthesis | IX |
| FISH (fluorescence in situ hybridization) | IV |
| FTPase | III |
| Gene knock-in experiments | VII |
| Gene knock-out experiments | VII |
| Helix-turn-helix | V |
| Homologous recombination | VII |
| Hox II | IX |
| HPLC | IV |
| In situ hybridization | IV |
| Initiation codon | V |
| Initiation complex | V |
| Interferon regulatory factor | X |
| Introns | V |
| IRF-1 | X |
| IRF-2 | X |
| Isoschizomer | III |
| Kinases | III |
| Klenow fragment | III |
| KOZAK sequence | V |
| LCR | V |
| Leucine zipper proteins | V |
| Library screening | IX |
| Ligases | III |
| Linkering | IX |
| Liposomes | VI |
| Locus control region | V |
| Long terminal repeat | VIII |
| Luciferase | V |
| Mammalian protein kinases | III |
| Master switch genes | V |
| Max | X |
| Maxam-Gilbert sequencing | IV |
| Minimal residual disease | IV |
| Missense mutation | V |
| MLL | X |
| Mobility shift (or band shift) assays | IV |
| mRNA | II |
| Mutagenesis, site-specific | IV |
| Nested PCR | IV |
| NF-1 | X |
| Nick-translation | IV |
| Nonsense mutation | V |
| Nonviral transduction methods | VIII |
| Northern blotting | IV |
| Nucleases | III |
| Nucleosomes | V |
| ORF (open reading frame) | II |
| p53 | X |
| PCR (polymerase chain reaction) | IV |
| Phage | II |
| Plasmids | II |
| Polyadenylation | V |
| Polylysine-ligand DNA | IX |
| Polymerases | III |
| Positional variegation | VIII |
| Post transcriptional regulation | V |
| Protein translation | V |
| Pseudotype retroviral vectors | IV |
| Pseudotyped viruses | VIII |
| Random priming | IV |
| RAR | X |
| Rb | X |
| RDA (representational difference analysis) | IX |
| Real-time PCR | IV |
| Reporter genes | V |
| Restriction endonuclease | III |
| Restriction fragment length polymorphism | XI |
| Retinoic acid receptor | X |
| Retroviral vectors | VIII |
| Reverse allele-specific hybridization | XI |
| Reverse genetics | IX |
| Reverse PCR | IV |
| Reverse transcriptase | III |
| RFLP | XI |
| Ribonuclease | III |
| Riboprobes | IV |
| Ribozyme | III |
| RNA (Ribonucleic acid) | II |
| RNA polymerase II | III |
| RNA polymerase III | III |
| RNAse protection assay | IV |
| S1 nuclease analysis | IV |
| SCL | X |
| Second strand synthesis | IX |
| Silencer | V |
| Southern blotting | IV |
| Southwestern blotting | IV |
| Splicing | V |
| Subtractive library | IX |
| Tal-1 | X |
| TATA | V |
| Tel | X |
| Telomere | II |
| Telomerase | III |
| Terminal deoxynucleotidyl | III |
| Thermostabile polymerases | III |
| Topoisomerase | III |
| Trans-acting factors | V |
| Transcription | V |
| Transcription factors | V |
| Transcriptional regulation | V |
| Transduction | VI |
| Transfection | VI |
| Transgenic animals | VIII |
| Transposon | VII |
| tRNA | II |
| Viral-derived kinases | III |
| Viral-derived transduction vectors | VIII |
| Western blotting | IV |
| X-linked methylation patterns | XI |
| YAC | VII |
| Yeast artificial chromosome | VII |
| Zinc finger domain proteins | V |
II. Nucleic Acids
DNA (deoxyribonucleic acid) The polymer constructed of successive nucleotides linked by phosphodiester bonds. Some 3 × 109 nucleotides are contained in the human haploid genome. During interphase, DNA exists in a nucleoprotein complex containing roughly equal amounts of histones and DNA, which interacts with nuclear matrix proteins. This complex is folded into a basic structure termed a nucleosome containing approximately 150 base pairs. From this highly ordered structure, DNA replication requires a complex process of nicking, unfolding, replication, and splicing. In contrast, gene transcription requires nucleosomal re-organization such that sites critical for the binding of transcriptional machinery reside at internucleosomal junctions.
Branched chain DNA (b-DNA) A method that exploits the formation of branched DNA to provide a sensitive and specific assay for viral RNA or DNA. The assay is performed in a microtiter format, in which partially homologous oligodeoxynucleotides bind to target to create a branched DNA. Enzyme-labeled probes are then bound to the branched DNA, and light output from a chemiluminescence substrate is directly proportional to the amount of starting target RNA. Standards provide quantitation. The assay displays a 4 log dynamic range of detection, with greater sensitivity to changes in viral load than RT-PCR-based assays. It has been employed to quantitate levels of HIV, HCV, and HBV.
RNA (ribonucleic acid) Three varieties of RNA are easily identified in the mammalian cell. Most abundant is ribosomal RNA (rRNA), which occurs in two sizes, 28S (approximately 4600 nucleotides) and 18S (approximately 1800 nucleotides); together they from the basic core of the eukaryotic ribosome. Messenger RNA (mRNA) is the term used to describe the mature form of the primary RNA transcript of the individual gene once it has been processed to eliminate introns and to contain a polyadenylated tail. mRNA links the coding sequence present in the gene to the ribosome, where it is translated into a polypeptide sequence. Transfer RNA (tRNA) is the form of RNA used to shuttle successive amino acids to the growing polypeptide chain. A tRNA molecule contains an anti-codon, a three-nucleotide sequence by which the tRNA molecule recognizes the codon contained in the mRNA template, and an adapter onto which the amino acid is attached.
Codon Three successive nucleotides on an mRNA that encode a specific amino acid in the polypeptide. Sixty-one codons encode the 20 amino acids, leading to codon redundancy, and three codons signal termination of polypeptide synthesis.
ORF (open reading frame) The term given to any stretch of a chromosome that could encode a polypeptide sequence, i.e., the region between a methionine codon (ATG) that could serve to initiate proteins translation, and the inframe stop codon downstream of it. Several features of the ORF can be used to judge whether it actually encodes an expressed protein, including its length, the presence of a “Kozak” sequence upstream of the ATG (implying a ribosome might actually bind there and initiate protein translation), whether the ORF exists within the coding region of another gene, the presence of exon/intron boundary sequences and their splicing signals, and the presence of upstream sequences that could regulate expression of the putative gene.
Plasmids Autonomously replicating circular DNA that are passed epigenetically between bacteria or yeast. In order to propagate, plasmids must contain an origin of replication. Naturally occurring plasmids transfer genetic information between hosts; of these, the genes encoding resistance to a number of antibiotics are the most important clinically. The essential components of plasmids are used by investigators to introduce genes into bacteria and yeast and to generate large amounts of DNA for manipulation.
Phage A virus of bacteria, phage such as lambda have been used to introduce foreign DNA into bacteria. Because of its infectious nature, the transfection (introduction) efficiency into the bacterial host is usually two orders of magnitude greater for phage over that of plasmids.
Cosmid By combining the elements of phage and plasmids, vectors can be constructed that carry up to 45 kb of foreign DNA.
cDNA A complementary copy of a stretch of DNA produced by recombinant DNA technology. Usually, cDNA represents the mRNA of a given gene of interest.
Telomere A repeating structure found at the end of chromosomes, serving to prevent recombination with free-ended DNA. Telomeres of sufficient length are required to maintain genetic integrity, and they are maintained by telomerase.
CpG This under-represented (i.e. < 1/16 frequency) dinucleotide pair is a “hotspot” for point mutation. CpG dinucleotides are often methylated on cytosine. Should Me-C undergo spontaneous deamination, uracil arises, which is then repaired by cellular surveillance mechanisms and altered to thymidine. The net result is a C to T mutation.
III. Enzymes of Recombinant DNA Technology
A. Nucleases
A number of common tools of recombinant DNA technology have been developed from the study of the basic enzymology of bacteria and bacteriophage. For example, most unicellular organisms have defense systems to protect against the invasion of foreign DNA. Usually, they specifically methylate their own DNA and then express restriction endonucleases to degrade any DNA not appropriately modified. From such systems come very useful tools. Today, most restriction endonucleases (and most other enzymes of commercial use) are highly purified from either natural or recombinant sources and are highly reliable. Using these tools, the manipulation of DNA and RNA has become routine practice in multiple disciplines of science.
Exonuclease An enzyme that digests nucleic acids starting from the 5′ or 3′ terminus and extending inward.
Endonuclease An enzyme that digests nucleic acids from within the sequence. Usually, specific sequences are recognized at the site where digestion begins.
Isoschizomer Restriction endonucleases that contain an identical recognition site but are derived from different species of bacteria (and hence have different names).
Restriction endonuclease These enzymes are among the most useful in recombinant DNA technology, capable of introducing a single cleavage site into a nucleic acid. The site of cleavage is dependent on sequence; recognition sites contain from 4 to 10 specific nucleotides. The resultant digested ends of the nucleic acid chain may either be blunt or contain a 5′ or 3′ overhang ranging from 1 to 8 nucleotides.
Ribonuclease These enzymes degrade RNA and exist as either exonucleases or endonucleases. The three most commonly used ribonucleases are termed RNase A, RNase T1, and RNAse H (which degrades duplex RNA or the RNA portion of DNA•RNA hybrids).
Ribozyme A complex RNA molecule that possesses endoribonuclease activity. A highly specific RNA sequence can generate secondary structure by virtue of intrachain base pairing. “Hairpin loops” and “hammer head” structures serve as examples of such phenomena. When the proper secondary structure forms, such RNA molecules can bind a second RNA molecule (e.g. an mRNA) at a specific location (dependent on an approximately 20-nucleotide recognition sequence) and cleave at a specific GUX triplet (where X = C, A, or U). These molecules will likely find widespread use as tools for specific gene regulation or as antiviral agents but are evolutionarily related to RNA splicing, which in its simplest form is autocatalytic.
B. Polymerases
DNA polymerase The enzyme that synthesizes DNA from a DNA template. The intact enzyme purified from bacteria (termed the holoenzyme) has both synthetic and editing functions. The editing function results from nuclease activity.
Klenow fragment A modified version of bacterial DNA polymerase that has been modified so that only the polymerase function remains; the 5′→3′ exonuclease activity has been eliminated.
Thermostabile polymerases The prototype polymerase, Taq, and newer versions such as Vent and Tth polymerase are derived from microorganisms that normally reside at high temperature. Consequently, their DNA polymerase enzymes are quite stable to heat denaturation, making them ideal enzymes for use in the polymerase chain reaction.
RNA polymerase II This enzyme is used by mammalian cells to transcribe structural genes that result in mRNA. The enzyme interacts with a number of other proteins to correctly initiate transcription, including a number of general factors, and tissue-specific and induction-specific enhancing proteins.
RNA polymerase III This enzyme is used by the cell to transcribe ribosomal RNA genes.
Kinases These enzymes transfer the γ-phosphate group from ATP to the 5′ hydroxyl group of a nucleic acid chain.
Viral-derived kinases These enzymes are utilized in recombinant DNA technology to transfer phosphate groups (either unlabeled or 32P-labeled) to oligonucleotides or DNA fragments. The most commonly used kinase is T4 polynucleotide kinase.
Mammalian protein kinases These enzymes transfer phosphate groups from ATP to either tyrosine, threonine, or serine residues of proteins. These enzymes are among the most important signaling molecules present in mammalian cell biology.
Farnesyl protein transferase (FTPase) FTPase adds 15 carbon farnesyl groups to CAAX motifs, such as one present in ras, allowing their insertion into cellular membranes.
Terminal deoxynucleotidyl This lymphocyte-specific enzyme normally transfers available (random) nucleotides to the 3′ end of a growing nucleic acid chain. In recombinant DNA technology, these enzymes can be used to add a homogeneous tail to a piece of DNA, thereby allowing its specific recognition in PCR reactions or in cloning efforts.
Ligases These enzymes utilize the g-phosphate group of ATP for energy to form a phosphodiester linkage between two pieces of DNA. The nucleotide contributing the 5′ hydroxyl group to the linkage must contain a phosphate, which is then linked to the 3′ hydroxyl group of the growing chain.
DNA methylases These enzymes are normally part of a bacterial host defense against invasion by foreign DNA. The enzyme normally methylates endogenous (host) DNA and thereby renders it resistant to a series of endogenous restriction endonucleases. In recombinant DNA work, methylation finds use in cDNA cloning to prevent subsequent digestion by the analogous restriction endonuclease.
Reverse transcriptase This enzyme, first purified from retrovirus-infected cells, produces a cDNA copy from an mRNA molecule if first provided with an antisense primer (oligo dT or a random primer). This enzyme is critical for converting mRNA into cDNA for purposes of cloning, PCR amplification, or the production of specific probes.
Topoisomerase A homodimeric chromosomal unwinding enzyme that introduces a double-stranded nick in DNA, which allows the unwinding necessary to permit DNA replication, followed by religation. Inhibition of topoisomerases leads to blockade of cell division, the target of several chemotherapeutic agents (e.g., etoposide).
Telomerase A specialized DNA polymerase that protects the length of the terminal segment of a chromosome. Should the telomere become sufficiently shortened (by repeated rounds of cell division), the cell undergoes apoptosis. The holoenzyme contains both a polymerase and an RNA template; only the latter has been characterized, although the gene for the enzymatic activity has recently been cloned.
IV. Molecular Methods
A number of molecular techniques have found widespread application in the biomedical sciences. This section of the glossary provides general concepts and is not intended to convey adequate details. The interested reader is referred to the excellent handbook of J. Sambrook and co-workers (Molecular Cloning, A Laboratory Manual, 2nd Ed., CSH Laboratory Press, 1989).
Maxam-Gilbert sequencing A method to determine the sequence of a stretch of DNA based on its differential cleavage pattern in the presence of different chemical exposures. A nucleic acid chain can be cleaved following G, A, C, or C and T by exposure of 32P-labeled DNA to neutral dimethylsulfate, dimethylsulfate-acid, hydrazine-NaCl-piperidine or hydrazine-piperidine alone, respectively.
Dideoxynucleotide (ddN) chain termination sequencing Also termed “Sanger sequencing,” this method relies on the random incorporation of dideoxynucleotides into a growing enzyme-catalyzed DNA chain. As no 3′ hydroxyl group is present on the ddN, chain synthesis halts following its incorporation into the chain. If 32P or 35S nucleotides are also incorporated into the reaction, a family of DNA fragments will be generated that can be visualized on a polyacrylamide gel. This method is presently the most commonly used chemistry to determine the sequence of DNA.
DNAse footprinting This technique depends on the ability of protein specifically bound to DNA to block the activity of the endonuclease DNAse I. 32P-labeled DNA is mixed with nuclear proteins, which potentially contain specific DNA-binding proteins, and the reaction is then subjected to limited DNAse digestion. If a given site of DNA is free of protein, it will be cleaved by the DNAse. In contrast, regions of DNAse specifically bound by proteins (transcription factors or enhancers) will be protected from digestion. The resultant mixture of DNA fragments from control and protein-containing reactions are then separated on a polyacrylamide gel. As the site of 32P labeling of the original DNA fragment is known, sites that were protected from DNAse digestion will be represented on the gel as a region devoid of that length fragment. Therefore, in comparison to naked DNA, regions that bind specific proteins will be represented as a “footprint.”
DNAse hypersensitivity site mapping This technique is designed to uncover regions of DNA that are in an “active” transcriptional state. It depends on the hypersensitivity of such sites (because of the lack of the highly compact nucleosome structure) to limited digestion with DNAse. Intact nuclei are subjected to limited DNAse digestion. The resultant large DNA fragments are then extracted, electrophoretically separated, and hybridized with a 32P-labeled probe from a known site within the gene of interest. If, for example, the probe were located at the site of transcription initiation, and should DNA fragments of 2 kb and 5 kb be detected with this probe, hypersensitive sites would thereby be mapped to 2kb and 5 kb upstream of the start of transcription initiation. By extrapolation, these sites would then be assumed important in the transcriptional regulation of the gene of interest, especially if such a footprint were only detected using cells that express that gene.
Mobility shift (or band shift) assays Like DNAse footprinting, this technique is also utilized to determine whether a fragment of DNA binds specific proteins. 32P-labeled DNA (either duplex oligonucleotides or small restriction fragments) are incubated with nuclear protein extracts and subjected to native acrylamide gel electrophoresis. Should specific DNA-binding proteins that recognize the oligonucleotide or restriction fragment probe be present in the nuclear extracts, a DNA-protein complex will be formed and its migration through the native gel will be retarded compared to the unbound DNA. Hence, the labeled band will be shifted to a more slowly migrating position. The specificity of their reaction can be demonstrated by also incubating, in separate reactions, competitor DNA that contains the presumed binding site or irrelevant DNA sequence.
S1 nuclease analysis This technique is used to identify the start of RNA transcription. The DNAse enzyme S1 cleaves only at sites of single-stranded DNA. Therefore, if 32P-labeled DNA is hybridized with mRNA, the resulting heteroduplex can be digested with S1, and the resulting DNA fragment will be of length equivalent to the site at which the piece of DNA begins through the mature 5′ end of the RNA.
RNAse protection assay This assay is in many ways similar to the S1 nuclease analysis. In this case, a 35S- or 32P-labeled antisense RNA probe is synthesized and hybridized with mRNA of interest. The duplex RNA is then subjected to digestion with RNAse A and T1, both of which will cleave only single-stranded RNA. Following digestion, the remaining labeled RNA is size-fractionated, and the size of the protected RNA probe then gives an indication of the size of the mRNA present in the original sample. This assay can also be used to quantitate the amount of specific RNA in the original sample.
PCR (polymerase chain reaction) This technique finds use in several arenas of recombinant DNA technology. It is based on the ability of sense and antisense DNA primers to hybridize to a cDNA of interest. Following extension from the primers on the cDNA template by DNA polymerase, the reaction is heat-denatured and allowed to anneal with the primers once again. Another round of extension leads to a multiplicative increase in DNA products. Therefore, a minute amount of cDNA can be efficiently amplified in an exponential fashion to result in easily manipulable amounts of cDNA. By including critical controls, the technique can be made quantitative. Important clinical examples of the use of PCR or reverse transcription PCR (see below) include (1) detection of diagnostic chromosomal rearrangements [e.g., bcr/abl in CML, t(15;17) in AML-M3, t(8;21) in AML-M2, or bcl-2 in follicular small cleaved cell lymphoma], or (2) detection of minimal residual disease following treatment. The level of sensitivity is one in 104 to 105 cells.
RT-PCR (reverse transcription PCR) This technique allows the rapid amplification of cDNA starting with RNA. The first step of the reaction is to reverse-transcribe the RNA into a first strand cDNA copy using the enzyme reverse transcriptase. The primer for the reverse transcription can either be oligo dT, to hybridize to the polyadenylation tail, or the antisense primer that will be used in the subsequent PCR reaction. Following this first step, standard PCR is then performed to rapidly amplify large amounts of cDNA from the reverse transcribed RNA.
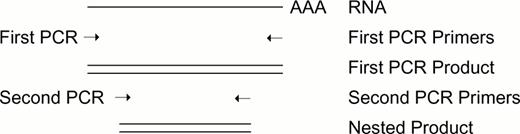
Nested PCR By using an independent set of PCR primers located within the sequence amplified by the primary set, the specificity of a PCR reaction can be greatly enhanced. In Figure 1, should the first PCR reaction yield a product of 600 nucleotides, a second PCR reaction using the first product as template and a different set of primers will produce a smaller, “nested” PCR product, the presence of which acts to confirm the identity of the primary product.
Real-time automated PCR During PCR, a fluorogenic probe, consisting of an oligodeoxynucleotide with both reporter and quencher dyes attached, anneals between the two standard PCR primers. When the probe is cleaved during the next PCR cycle, the reporter is separated from the quencher so that the fluorescence at the end of PCR is a direct measure of the amplicons generated throughout the reaction. Such a system is amenable to automation and gives precise quantitative information.
Allele-specific PCR By using generic PCR primers flanking the immunoglobulin or T cell receptor genes, the precise rearranged gene characteristic of a B or T cell neoplasm can be amplified and sequenced. Once so obtained, new PCR primers can then be designed that are unique to the patient's tumor. Such allele-specific PCR can then be used to detect blood cell contamination by tumor and to detect minimal residual disease following therapy.
Southern blotting This technique is used to detect specific sequences within mixtures of DNA. DNA is size-fractionated by gel electrophoresis and then transferred by capillary action to nitrocellulose or another suitable synthetic membrane. Following blocking of nonspecific binding sites, the nitrocellulose replica of the original gel electrophoresis experiment is then allowed to hybridize with a cDNA or oligonucleotide probe representing the specific DNA sequence of interest. Should specific DNA be present on the blot, it will combine with the labeled probe and be detectable by autoradiography. By co-electrophoresing DNA fragments of known molecular weight, the size(s) of the hybridizing band(s) can then be determined. For gene rearrangement studies, Southern blotting is capable of detecting clonal populations that represent approximately 1% of the total cellular sample.
Northern blotting This modification of a Southern blot is used to detect specific RNA. The sample to be size-fractionated in this case is RNA and, with the exception of denaturation conditions (alkali treatment of the Southern blot versus formamide/formaldehyde treatment of the RNA sample for Northern blot), the techniques are essentially identical. The probe for Northern blotting must be antisense.
Western blotting This technique is designed to detect specific protein present in a heterogenous sample. Proteins are denatured and size-fractionated by polyacrylamide gel electrophoresis, transferred to nitrocellulose or other synthetic membranes, and then probed with an antibody to the protein of interest. The immune complexes present on the blot are then detected using a labeled second antibody (for example, a 125I-labeled or biotinylated goat anti-rabbit IgG). As the original gel electrophoresis was done under denaturing and reducing conditions, the precise size of thee target protein can be determined.
Southwestern blotting This technique is designed to detect specific DNA-binding proteins. Like the Western blot, proteins are size-fractionated and transferred to nitrocellulose. The probe in this case, however, is a double-stranded labeled DNA that contains a putative protein-binding site. Should the DNA probe hybridize to a specific protein on the blot, that protein can be subsequently identified by autoradiography. This technique often suffers from nonspecificity, so that a number of critical controls must be included in the experiment for the results to be considered rigorous.
In situ hybridization This technique is designed to detect specific RNA present in histological samples. Tissue is prepared with particular care not to degrade RNA. The cells are fixed on a microscope slide, allowed to hybridize to probe, and then washed and overlaid with photographic emulsion. Following exposure for one to four weeks, the emulsion is developed and silver grains overlying cells that contain specific RNA are detected. The most useful probes for this purpose are metabolically 35S-labeled riboprobes generated by in vitro transcription of a cDNA using viral RNA polymerase. These probes give the lowest background and are preferable to using terminal deoxynucleotidyl transferase or alternative methods using 32P as an isotope.
FISH (fluorescence in situ hybridization) A general method to assign chromosomal location, gene copy number (both increased and decreased), or chromosomal rearrangements. Biotin-containing nucleotides are incorporated into specific cDNA probes by nick-translation. Alternatively, digoxigenin or fluorescent dyes can be incorporated by enzymatic or chemical methods. The probes are then hybridized with solubilized, fixed metaphase cells, and the copy number of specific chromosomes or genes are determined by counter-staining with fluorescein isothiocyanate (FITC)-labeled avidin or other detector reagents. The number and location of detected fluorescent spots correlates with gene copy number and chromosomal location. The method also allows chromosomal analysis in interphase cells, allowing extension to conditions of low cell proliferation.
CGH (comparative genome hybridization) In CGH, DNA is extracted from tumor and from normal tissues and differentially labeled with fluorescent dyes. Once the DNA samples are mixed and hybridized to normal metaphase chromosome spreads, chromosomal regions that are under-represented or over-represented in the tumor sample can be identified. This method can be applied to extremely small tumor samples (by using PCR methods) of formalin-fixed or frozen tissue. It has been applied to detect loss of chromosome 18q or 17p in colon cancer and is likely to be applied to hematologic malignancies. The sensitivity of the technique approaches 1 cell in 100.
Nick-translation This technique is used to label cDNA to high specific activity for the purpose of probing Southern and Northern blots and screening cDNA libraries. The cDNA fragment is first nicked with a limiting concentration of DNAse, then DNA polymerase is used to both digest and fill in the resulting gaps with labeled nucleotides.
Random priming This technique is also used to produce labeled cDNA probes and is dependent on using random 6- to 10-base oligonucleotides to sit down on a single-stranded cDNA and then using DNA polymerase to synthesize the complementary strand using labeled nucleotides. This technique usually produces more favorable results than nick-translation.
Riboprobes These labeled RNA molecules are produced by first cloning the cDNA of interest into a plasmid vector that contains promoters for viral RNA polymerases. Following cloning, the viral RNA polymerase is added, and labeled nucleotides are incorporated into the resulting RNA transcript. This molecule is then purified and used in probing reactions. Many such cloning vectors (for example, pGEM) have different RNA polymerase promoters on either side of the cloning site, allowing the generation of both sense and antisense probes from the same construct.
Mutagenesis, site-specific Several methods are now available to intentionally introduce specific mutations into a cDNA sequence of interest. Most are based on designing an oligonucleotide that contains the desired mutation in the context of normal sequence. This oligonucleotide is then incorporated into the cDNA using DNA polymerase, either using a single-stranded DNA template (phage M13) or in a PCR format to produce a heteroduplex DNA containing both wild type and mutant sequences. Using M13, recombinant phage are then produced and mutant cDNA are screened for on the basis of the difference in wild type and mutant sequences; using the PCR format, the exponential amplification of the mutant sequence results in its overwhelming numerical advantage over wild type sequence, resulting in nearly all clones containing mutant sequence. Both of these methods require that the entire cDNA insert synthesized in vitro be sequenced in its entirety to guarantee the fidelity of mutagenesis and synthesis of the remaining wild type sequences.
Chromatography, gel filtration This technique is designed to separate proteins based on their molecular weight. It is dependent on the exclusion of proteins from a matrix of specific size. Proteins that are too large to fit into the matrix of the gel bed run to the bottom of the column more quickly than smaller proteins, which are included in the volume of the matrix. Therefore, using appropriate size markers, the approximate molecular weight of a given protein can be determined and it can be separated from proteins of dissimilar size. Typical separation media for gel filtration chromatography include Sephadex and Ultragel.
Chromatography, ion exchange This separation methodology depends on the preferential binding of positively charged proteins to a matrix containing negatively charged groups or a negatively charged protein binding to a matrix containing positively charged groups. Increases in the buffer concentration of sodium chloride are then used to break the ionic interaction between protein and matrix and elute off-bound proteins. Examples of such separation media include DEAE and CM cellulose.
Chromatography, hydrophobic This methodology separates proteins based on their hydrophobicity. Proteins preferentially bind to the matrix based on the strength of this interaction; proteins are then eluted off using solvents of increasing hydrophobicity. Separation media include phenyl-sepharose and octyl-sepharose.
Chromatography, affinity This separation method depends on using any molecule that can preferentially bind to a protein of interest. Typical methodologies include using lectins (such as wheat germ or concanavalin A) to bind glycoproteins or using covalently coupled monoclonal antibodies to bind specific protein ligands.
Chromatography, high performance liquid (HPLC)
A general methodology to improve the separation of complex protein mixtures. The types of HPLC columns available are the same as for conventional chromatography, such as those based on size exclusion, hydrophobicity, and ionic interaction, but the improved flow rates resulting from the high pressure system provide enhanced separation capacity and improved speed.
V. Physiologic Gene Regulation
The regulation of gene expression is central to physiology. Complex organisms have evolved multiple mechanisms to accomplish this task. The first step in protein expression is the transcription of a specified gene. The rate of initiation and elongation of this process is the most commonly used mechanism for regulating gene expression. Once formed, the primary transcript must be spliced, polyadenylated, and transported to the cytoplasm. These mechanisms are also possible points of regulation. In the cytoplasm, mRNA can be rapidly degraded or retained, another potential site of control. Protein translation next occurs on the ribosome, which can be free or membrane-associated. Secreted proteins take the latter course, and the trafficking of the protein through these membranes and ultimately to storage or release makes up another important point of potential regulation. Individual gene expression is often controlled at multiple levels, making investigation and intervention a complex task.
Transcription Transcription is the act of generating a primary RNA molecule from the double-stranded DNA gene. Regulation of gene expression is predominantly at the level of regulating the initiation and elongation of transcription. The enzyme RNA polymerase is the key feature of the system, which acts to generate the RNA copy of the gene in combination with a number of important proteins. There is usually a fixed start to transcription and a fixed ending.
TATA Many genes have a sequence that includes this tetranucleotide close to the beginning of gene transcription. RNA polymerase binds to the sequence and begins transcription at the cap site, usually located approximately 30 nucleotides downstream.
Enhancer An enhancer is a segment of DNA that lies either upstream, within, or downstream of a structural gene that serves to increase transcription initiation from that gene. A classical enhancer element can operate in either orientation and can operate up to 50 kb or more from the gene of interest. Enhancers are cis-acting in that they must lie on the same chromatin strand as the structural gene undergoing transcription. These cis-acting sequences function by binding specific proteins, which then interact with the RNA polymerase complex.
Silencer These elements are very similar to enhancers except that they have the function of binding proteins and inhibiting transcription.
Initiation complex This multi-protein complex forms at the site of transcription initiation and is composed of RNA polymerase, a series of ubiquitous transcription factors (TF II family), and specific enhancers and/or silencers. The proteins are brought together by the looping of DNA strands so that protein binding sites, which may range up to tens of kb apart, can be brought into close juxtaposition. Specific protein•protein interactions then allow assembly of the complex.
Polyadenylation Following transcription of a gene, a specific signal near the 3′ end of the primary transcript (AATAAA) signals that a polyadenine tail be added to the newly formed transcript. The tail may be up to several hundred nucleotides long. The precise function of the poly A tail is uncertain but it seems to play a role in stability of the mRNA and perhaps in its metabolism through the nuclear membrane to the ribosome.
Splicing The primary RNA transcript contains a number of sequences that are not part of the mature mRNA. These regions are called introns and are removed from the primary RNA transcript by a process termed splicing. A complex tertiary structure termed a lariat is formed and the intron sequence is eliminated bringing the coding sequences (exons) together. Specific sequences within the primary transcript dictate the sites of intron removal.
Exons These are the regions of the primary RNA transcript that, following splicing, form the mature mRNA species, which encodes polypeptide sequence.
Introns These are the regions of the primary RNA transcript that are eliminated during splicing. Their precise function is uncertain. However, several transcriptional regulatory regions have been mapped to introns, and they are postulated to play an important role in the generation of genetic diversity (exon shuffling mechanism).
Nucleosomes When linear, the length of a specific chromosome is many orders of magnitude greater than the diameter of the nucleus. Therefore, a mechanism must exist for folding DNA into a compact form in the interphase nucleus. Nucleosomes are complex DNA protein polymers in which the protein acts as a scaffold around which DNA is folded. The mature chromosomal structure then appears as beads on a string; within each bead (nucleosome) are folded DNA and protein. Nucleosome structure is quite fluid, and internucleosomal stretches of DNA are thought to be sites that are important for active gene transcription.
Trans-acting factors Proteins that are involved in the transcriptional regulation of a gene of interest.
Cis-acting factors These are regions at a gene either upstream, within, or downstream of the coding sequence that contains sites to which transcriptionally important proteins may bind. Sequences that contain 5 to 25 nucleotides are present in a typical cis-acting element.
Transcription factors Specific proteins that bind to control elements of genes. Several families of transcription factors have been identified and include helix-loop-helix proteins, helix-turn-helix proteins, and leucine zipper proteins. Each protein includes several distinct domains such as activation and DNA-binding regions.
LCR (locus control region)Cis-acting sites are occasionally organized into a region removed from the structural gene(s) they control. Such locus control regions (LCRs) are best described for the b globin and a globin loci. First recognized by virtue of clustering of multiple DNAse hypersensitive sites, the β globin LCR is required for high level expression from all of the genes and appears to be critical for their stage-specific developmental pattern of expression.
Protein translation This term is applied to the assembly of a polypeptide sequence from mRNA.
KOZAK sequence This five-nucleotide sequence resides just prior to the initiation codon and is thought to represent a ribosomal-binding site. The most consistent position is located three nucleotides upstream from the initiation ATG and is almost always an adenine nucleotide. When multiple potential initiation codons are present in an open reading frame, the ATG codon, which contains a strong consensus KOZAK sequence, is likely the true initiation codon.
Initiation codon The ATG triplet is used to begin polypeptide synthesis. This is usually the first ATG codon, located approximately 30 nucleotides downstream of the site of transcription initiation (cap site). However, the context in which the ATG resides is also important (see KOZAK sequence).
Missense mutation Mutation of the mRNA sequence to generate an altered codon, which results in an amino acid change, is termed a missense mutation.
Nonsense mutation This type of mutation results in the generation of a premature termination codon and hence creates a truncated polypeptide.
Transcriptional regulation Gene regulation is determined by the rate of transcriptional initiation. This usually results from alteration in the level of activity of transacting proteins, which, in turn, are regulated either by the amount of the transcriptionally active protein or by their state of activation.
Leucine zipper proteins A family of DNA-binding proteins that require a dimeric state for activity and that dimerize by virtue of an alpha helical region that contains leucine at every seventh position. Because 3.4 amino acids reside in each turn of an alpha helix, the occurrence of leucine at every seventh position results in a strip of highly hydrophobic residues on one surface of the alpha helix. Such a domain on one polypeptide can intercollate with a similar domain on a second polypeptide, resulting in the formation of a stable homodimer or heterodimer. Examples of the leucine zipper family include the proto-oncogenes c-jun and c-fos.
Basic helix-loop-helix proteins These transcriptional proteins are characterized by two alpha helical regions separated by a loop structure; this domain is involved in protein dimerization. Examples of this family of transcription factors include E12/E47 of the immunoglobulin promoter or Myo D of muscle cell regulation.
Helix-turn-helix This family of transcriptionally active proteins depends on the helix-turn-helix motif for dimerization. Examples include the homeodomain genes such as the Hox family.
Master switch genes These polypeptide products are thought to regulate a whole family of genes and result in a cell undergoing a new program of differentiation. An example of such a system is Myo D, in which activation is thought to lead to differentiation along the muscle cell lineage.
Zinc finger domain proteins The presence of conserved histidine and cysteine residues allows chelation of a zinc atom and results in the formation of a loop structure called the zinc finger domain. This feature is present in a large family of transcriptionally active proteins such as the steroid hormone receptors.
Post-transcriptional regulation Mechanisms of gene regulation that do not involve transcriptional enhancement or silencing and include altering the rate of mRNA degradation, the efficiency of translation or post-translational modification, or transportation of the polypeptide out of the cell.
Actinomycin D pulse experiments The application of actinomycin D to actively metabolizing cells results in the cessation of new RNA transcription. Consequently, serial determinations of specific RNA levels will allow one to calculate the mRNA half life. Should this vary between control and stimulated conditions, evidence is garnered that a gene of interest is regulated at the level of mRNA stability.
Reporter genes In order to determine how a gene promoter or enhancer works in vitro, that genetic element is often linked to a gene for which a simple assay is readily available and whose regulation is not affected by post-transcriptional processes. Such reporter genes include chloramphenicol acetyl transferase, β galactosidase, and firefly luciferase. The first is the most commonly used reporter; however, more recent studies have emphasized the use of the latter two reporters, as these are more sensitive to minimal changes in promoter or enhancer activity.
CAT (chloramphenicol acetyl transferase) The bacterial gene for chloramphenicol resistance, chloramphenicol acetyl transferase (CAT) is commonly used as a reporter gene for investigating physiologic gene regulation. The assay depends on the ability of transfected cellular cytoplasm to convert 14C chloramphenicol to its acetylated form in the presence of acetyl CoA. The acetylated forms are separated from the 14C substrate using thin layer chromatography.
β galactosidase The presence of β galactosidase activity in the cytoplasm of transfected cells can be readily detected by its ability to convert a colorless substrate to a blue-colored product. This is usually assayed using a fluorimeter.
Luciferase This gene, which is the most recent reporter gene to be used, has gained increasing acceptance because of its ease of assay and extreme sensitivity. The assay is based on the ability of the protein to undergo chemiluminescence and transmit light, detected with a luminometer.
Cyclins A group of proteins that vary in expression throughout the cell cycle. Once a threshold level is attained, interaction with specific cellular kinases results in phosphorylation of critical components of the mitotic machinery. Several classes of cyclins (A through E) exist that regulate different aspects of the cell cycle (G0, G1, S, G2, M). Altered expression of some cyclins is associated with hematologic malignancy, e.g., t(11;14) in mantle cell lymphoma leads to over-expression of cyclin D1, a G1 phase cyclin.
Cdk (cyclin-dependent kinase) A related group of cellular kinases, present in virtually all cells, that are regulated both positively and negatively by specific phosphorylation events and negatively by association with other proteins, and are dependent on cyclins, present only during certain phases of the cell cycle (cdk1-activated during G2/M phase, cdk2-G1/S phases, cdk4-G1/S phases, cdk6-G1 phase, cdk7-throughout the cell cycle).
CdkI (cdk inhibitors) Proteins that inhibit the cdks by stoicheometric combination, arresting cells in G1 phase, and include p27, p21 and the p16 Ink 4A family of proteins. The latter are implicated as tumor suppressor genes, as their deficiency in mice leads to rapid cellular proliferation and a high rate of spontaneous tumor development. Moreover, deficiency of p16 family members has been associated with numerous types of human tumors, including a fraction of cases of B cell ALL and T cell leukemia.
VI. Expression of Recombinant Proteins
In order to exploit the techniques of recombinant DNA research, one must possess a system to manufacture the protein of interest. After identifying the gene encoding the protein and obtaining a cDNA representation of it (“cloning”), the cDNA must be placed in a vector capable of driving high levels of RNA transcription in a host system capable of translating and appropriately modifying the polypeptide to produce fully functional protein. And just like obtaining a protein of interest from natural sources, one must purify protein from the final expression system. Because of the nature of the highly engineered systems and high levels of expression, this latter task is usually considerably easier using recombinant methods than from natural sources. The methods used to generate expression vectors are described in Section IV, but the methods to purify proteins are discussed in only a rudimentary way and are beyond the scope of this glossary.
Expression vector A plasmid that contains all of the elements necessary to express an inserted cDNA in the host of interest. For a mammalian cell host, such a vector typically contains a powerful promoter coupled to an enhancer, a cloning site, and a polyadenylation signal. In addition, several expression vectors also contain a selectable marker gene such as DHFR or NeoR, which aids in the generation of stable cell lines. The plasmid also requires a bacterial origin of replication and an antibiotic resistance gene (AmpR) to allow propagation and expansion in a bacterial host.
Transfection Once the expression vector has been assembled, it must be inserted into the host of interest. Several methods are available for such transfections and include calcium/phosphate/DNA complexes, DEAE Dextran, electroporation, liposome, and retrovirus-mediated gene transfer.
Calcium phosphate This method relies on the production of a calcium/phosphate/DNA microprecipitate, which is then taken up by cells by pinocytosis. The method is very effective for a number of commonly used mammalian cell expression systems including COS, BHK, 293, and CHO cells.
DEAE dextran This method depends on the formation of a complex between the insoluble positively charged dextran and the DNA to be transfected. Like calcium phosphate, this method is highly successful with many cell types.
Electroporation When cells are suspended in buffer between two electrodes, discharge of an electrical impulse momentarily creates pores in the cell membrane. During this time, DNA in solution is free to diffuse into the cells. This method is highly successful in transfecting a large number of cell types, including cells previously thought to be difficult to transfect with other methods, such as endothelial cells and fibroblasts.
Liposomes By encapsulating the DNA to be transfected in an artificial lipid carrier, foreign DNA can be introduced into the cell. This method, like electroporation, has been successful in transfecting cells previously thought difficult to manipulate. Its only drawback is its expense.
Transduction The act of transferring a foreign gene into a host genome.
VII. Experimental Gene Manipulation
Antisense oligonucleotides By introducing short single-stranded deoxyribonucleic acids (ODN) into a cell, specific gene expression can be interrupted. Several mechanisms have been postulated to account for these results including interruption of ribosome binding to mRNA, enhanced degradation of mRNA mediated by the double-strand specific RNAseH, DNA triplex formation, and impairment of translation efficiency. Most successful attempts using antisense ODN have targeted sequences surrounding and including the initiation codon. To reduce nuclease attack, the antisense ODN are often synthesized using an altered chemistry involving thiol rather than phosphodiester linkages.
Transgenic animals By introducing an intact or manipulated gene into the germline of mice, the effects of promoter expression in specific cell lineages can be investigated. In contrast to highly artificial in vitro studies using reporter gene analysis, such transgenic animals provide an important in vivo model of gene function. The methods for production of transgenic mice have been extensively reviewed and are based on the microinjection of linear DNA into the pronucleus of a fertilized egg. Several types of experiments can be performed. First, the effect of aberrant expression of a gene can be investigated, as was recently performed by expressing GM-CSF in a wide variety of tissues. Second, the necessary elements for tissue- and developmental level-specific expression of a gene can be studied, as has been performed for the β-globin locus. Third, the tissue distribution of a specific gene can be determined by engineering a marker gene adjacent to a specific promoter. A specific example of this strategy employs a “suicide gene,” the herpes virus thymidine kinase (TK). When animals carrying such genes are exposed to gancyclovir, cells expressing the promoter of interest will express TK, be killed, and be readily detected.
Gene knock-out experiments Specific genes in the mammalian genome can now be targeted for interruption or correction based on the technique of homologous recombination. By generating DNA constructs that contain an interrupted gene of interest, or a corrected gene, in the setting of adequate flanking sequences to allow for targeting to the genetic locus of interest, the endogenous gene can be replaced or corrected. The methods involve introduction of the gene into an embryonic stem (ES) cell line, selection for subclones of cells that have had successful homologous recombination events, and then introduction of the ES subclone into the blastocyst of a developing embryo. A chimeric animal results, and should the newly introduced gene become part of the germline, it can be bred to the homozygous state. Using these techniques, investigators can now determine whether a single genetic locus is responsible for a given disease, determine the significance of specific cytokines or growth factors, and generate model systems useful investigation of human disease.
Gene knock-in experiments A similar technology to knock-out strategy, but rather than simply obliterating function of the targeted gene, the knock-in is designed to replace the locus with a specific mutation of interest.
Homologous recombination When a manipulated gene is introduced into a cell, it can be incorporated into the genome either randomly or at a specific locus. By incorporating sequences that normally flank the desired locus, a manipulated gene can be specifically (albeit rarely) introduced into the genome. Selection for this unlikely event can be enhanced by introduction of the herpes thymidine kinase (TK) gene into the original targeting construct. Should the construct be randomly incorporated into the genome, the TK gene will also be introduced, rendering the cell sensitive to gancyclovir. If homologous recombination occurs, the TK gene will be eliminated, as there are no homologous sequences at the specific genetic locus of interest and the resultant cell will be resistant to the antibiotic.
YAC (yeast artificial chromosome) A yeast artificial chromosome (YAC) utilizes centromeric and telomeric elements from yeast chromosomes to construct genetic elements that can be propagated in yeast and transferred into mammalian cells. Such vehicles allow the introduction of up to 200 kb or more of genetic material into the host cells. YACs are now being used to study the physiologic regulation of large genetic loci such as the β-globin region of chromosome 11.
Contig The jargon term used to describe the assembly of clones necessary to include all of the DNA in a specific stretch of chromosome. Such maps are usually assembled from overlapping YAC (yeast artificial chromosome) or BAC (bacterial artificial chromosome) clones. Once the “genome project” is complete, it will consist of 24 (very large) contigs (22 autosomal, an X and a Y).
Transposon Naturally occurring genetic elements that are naturally easily removed and inserted into the genome, allowing for the recombination of genetic segments, giving rise to genetic diversity. These same elements can be utilized for gene therapy.
VIII. Gene Therapy
Gene therapy takes many forms. To treat malignancy, it may involve the insertion of an adjuvant substance (such as GM-CSF) into tumor cells to generate a tumor vaccine, transfer of a gene that renders tumor cells susceptible to eradication with an antitumor agent (e.g., herpes thymidine kinase), or insertion of a gene that makes bystander cells resistant to the effects of chemotherapy (e.g., MDR). For gene deficiencies, insertion of the wild type allele is the therapeutic goal. Obtaining cDNA for desired genes has become common. Insertion of the gene into target cells and high (adequate) level expression is more problematic. Several types of transfer vehicles have found use, including viral vectors and chemical agents.
Viral transduction vectors Retroviral vectors are based on murine retroviruses. They can carry 6 to 7 kb of foreign DNA (promoter + cDNA) but suffer from the drawbacks of requiring the development of high titer packaging lines, requiring that target cells be dividing, and are subject to host cell down-modulation. Adenoviral vectors can be produced at high levels and do not require a dividing target cell, but they do not normally integrate, resulting in only transient expression. Adeno-associated viral vectors are defective parvoviruses that integrate into a non-dividing host cell at a specific location (19q). Disadvantages are genetic instability, small range of insert size (2-4.5 kb), and thus far, only transient expression.
Ecotropic vectors Many retroviruses are host cell specific, i.e. they will only infect a specific species of cells. An example is the widely used Maloney virus, and its basis lies in the species-specific expression of the viral cell surface receptor.
Ecotropic viruses Murine retroviruses that contain coat proteins that can only bind to murine cellular receptors.
Amphotropic viruses Retroviruses whose coat proteins bind to a receptor found throughout multiple species, usually including man, making these vectors suitable for human use. Problems related to the level of receptor expression on cells of hematologic interest (e.g. stem cells) remain for amphotropic viruses.
Pseudotyped virus These take advantage of the powerful expression levels obtainable by murine retroviral backbones, yet are packaged in an envelope that allows docking and uptake by human target cells. An example is the popular MFG vector that utilizes a murine leukemia retroviral backbone and an amphotropic packaging cell line to produce infectious particles.
Episomal Episomal refers to gene therapy vectors that remain free in the target cell without being taken into the host genome.
Positional variegation Refers to the observation that the site of vector integration into the genome often results in variable levels of gene expression.
Chimeraplasty A technique of gene therapy dependent on construction of a DNA:RNA oligonucleotide hybrid that once introduced into a cell relies upon DNA repair mechanisms to introduce a (corrective) change in the targeted gene.
Chitosan-DNA A chemical means of packaging foreign DNA to allow introduction into cells; the complexes exist as nanospheres and have been tested in factor IX deficiency in animals.
Long terminal repeat (LTR) This segment of a retroviral genome carries the genetic information for both transcription of downstream viral structural genes and the mechanisms of viral replication. It is often used in retroviral applications to drive the exogenous therapeutic gene as it carries a powerful (but non-tissue specific) promoter.
Interference The mechanisms by which infection of a cell by one virus excludes infection by others. Interference is often due to the cellular production of coat proteins, which bind to and block the cells' remaining viral receptors.
Nonviral transduction methods Nonviral methods include polylysine-ligand DNA complexes, where the ligand (e.g., transferrin) allows access to the cell through normal receptor-mediated uptake, and phospholipid vesicles. Both methods suffer from not providing a mechanism for genomic integration, precluding long-term expression.
IX. Cloning and Library Screening
Obtaining cDNA representing a protein of interest is usually the first step in the process of applying the techniques of recombinant DNA research to an important physiologic question. A suitable cDNA library must first be constructed starting with RNA abundant (or as abundant as possible) in the transcripts for the gene of interest. Following library construction a probe must be developed that can specifically recognize the gene or cDNA of interest, or the expressed protein product of the specific cDNA.
First strand synthesis The retroviral enzyme reverse transcriptase is used along with an antisense primer to produce a complementary DNA strand of mRNA extracted from a cellular source known to express the gene of interest. Two types of primers are used, either oligo dT, in which the poly A tail begins the cDNA synthesis, or random primers, in which a whole range of start sites will be used.
Second strand synthesis The enzyme DNA polymerase is used to generate the sense strand of cDNA. Priming of the second strand can occur spontaneously, as the anti-sense first cDNA strand can form a hairpin loop at its 3′ end bending back to prime second strand synthesis. Alternatively, a polynucleotide tail can be added to the first strand synthesis using terminal deoxynucleotide transferase, then second strand priming can occur using a synthetic oligonucleotide complementary to the TdT tail. Should the former technique be used, an extra step to nick the hairpin loop using the enzyme S1 nuclease would be required prior to inserting the cDNA into its vector.
cDNA blunting First and second strand synthesis usually results in nonflush ends. To prepare the cDNA for insertion into a cloning vector, the ends must be made flush with one another. Such blunting reactions can be conducted with a DNA polymerase, such as the Klenow fragment of DNA polymerase I or T4 DNA polymerase.
Linkering To efficiently insert the cDNA library into a cloning vector, synthetic duplex oligonucleotides that contain a restriction endonuclease site are attached to the blunted ends of the cDNA. A restriction endonuclease is chosen that rarely cuts DNA (such as the 8 bp recognition sequence for Not I, or if a more common restriction site is used such as Eco RI, the cDNA should first be methylated in order to prevent subsequent cDNA digestion with the enzyme) and is used to generate “sticky ends” on the cDNA.
cDNA library preparation Once the cDNA has been prepared and sticky ends generated, the library is inserted into a convenient cloning vector. Because of high cloning efficiency, most cDNA libraries are constructed in a λ phage vector. Typically, if screening is to be performed using a monoclonal antibody, λ gt 11 is used. If screening is to be performed using oligonucleotide probes, λ gt 10 can be used. If larger DNA fragments are to be prepared, such as from genomic fragments of DNA, λ vectors that can accommodate up to 20 kb are available (e.g., λ Charon 4A).
Subtractive library The purpose of generating a subtractive library is to enrich for cDNA that are expressed under one condition but are not expressed under a second condition. This facilitates screening for the cDNA of interest in that the complexity of the library is much reduced, requiring one to screen far fewer clones. At its extreme, investigators have used subtractive libraries to generate a very highly select group of clones (in the range of 100) and then have sequenced all of the resulting cDNA. The principle behind a subtractive library is the elimination of cDNA common to induced and control conditions. By eliminating such clones, only cDNA that are present under the induced conditions will remain in the library. Those techniques depend on the differential elimination of duplex mRNA/cDNA or cDNA/cDNA hybrids, which form between genes expressed under both conditions, leaving the single-stranded mRNA or cDNA of interest.
RDA (representational difference analysis) A molecular method to amplify genes that are expressed in an RNA sample of interest, that are not present, or present at very reduced levels, in a comparison RNA sample (e.g. cytokine induced and control cells). The method relies on RT-PCR amplification of the RNA that does not contain the gene(s) of interest to produce a “driver” cDNA, and RT-PCR to produce “tester” cDNA from the RNA population in which you hope to find new genes. After ligation of different oligonucleotides to the ends of each population, both are denatured and an excess of the driver is hybridized to the tester and PCR performed with primers that will amplify only sequences present in the tester that are not in the driver, thereby “removing” cDNA common to both populations. The resultant cDNA are enriched in uniquely expressed genes.
Directional cloning To improve efficiency when screening functional expression libraries, many investigators construct cDNA libraries in which the proper coding orientation of the cDNA is maintained in the library. In conventional library preparation, the 5′ and 3′ ends of the DNA are identical; thus, cDNA can be inserted into the cloning vector in either orientation. If screening is dependent on the production of a functional protein, one-half of the library will be useless, as those cDNA inserted in an inverse orientation will not produce functional protein. Directional cloning is dependent on producing sticky ends that differ on the 5′ and 3′ termini. The cloning vector has the appropriate pair of complementary cloning sites.
Library screening Three major methods are available to obtain cDNA of interest. The classic technique utilizes DNA probes (such as oligonucleotides or intact cDNA from a homologous gene) to screen cDNA libraries. An oligonucleotide probe is usually derived from a reverse translation of known protein sequence. By expressing cDNA as a fusion protein with β galactosidase, various antisera can be used to screen for fusion proteins encoded by the cDNA of interest. Finally, cDNA libraries may be constructed in cloning vectors that allow for expression of the cDNA insert in E. coli or a mammalian cell host. If a highly sensitive assay for the desired protein's function can be developed, pools of cDNA clones can be expressed and then assayed together; a positive assay from a pool would allow one to subdivide into smaller pools and eventually at clonal density.
Reverse genetics Often, large families of homologous proteins exist and multiple previously unknown members of the family can be obtained by screening cDNA libraries under low stringency using cDNA or oligonucleotide probes from regions highly conserved amongst members of the family. In this case, genes are identified before their function is known, a situation referred to as reverse genetics. Examples in hematology include identifying members of the tyrosine kinase family of receptor proteins using a probe derived from the conserved kinase domain of the cytoplasmic region of src or other tyrosine kinase proto-oncogenes, or the identification of transcription factors important in hematopoiesis using conserved motifs present in zinc finger or homeodomain proteins.
X. Oncogenesis and Anti-Oncogenes
Oncogenes have usually been identified in the context of a tumor-inducing virus. Such viral oncogenes (v-onc) are thought to be derived from host cells, but have been altered such that abnormal regulation of production or function has ensued during the transfer process. Subsequent reintroduction of the altered gene into a host cell leads to transformation. Proto-oncogenes, the normal cellular counterpart of viral oncogenes, can contribute to cellular transformation by mechanisms that disturb normal gene function. Such mechanisms include mutation (resulting in abnormal function), amplification (resulting in abnormal levels of expression), rearrangement (resulting in a new function), or promoter mutation (again resulting in abnormal levels of expression). Most or all protooncogenes are involved in normal cellular processes such as growth factor signal transduction, mitogenic signaling, or regulation of DNA transcription or cellular proliferation. The nomenclature convention is to indicate the cellular version of the proto-oncogene as “c-onc” and the viral version, which is transforming, as “v-onc.” Most altered proto-oncogenes act in a dominant genetic fashion. Anti-oncogenes, or tumor suppressor genes, usually act in a recessive genetic fashion and function to slow processes involved in cellular proliferation. Most of the identified anti-oncogenes have been involved in gene transcription, presumably acting to enhanced differentiation programs over those of proliferation.
c-abl This gene, present on human chromosome 9, encodes a tyrosine kinase whose role in normal hematopoiesis is unclear; however, its fusion to the BCR gene on human chromosome 22, the functional counterpart of the Ph1 chromosome strongly associated with the disease chronic myelogenous leukemia, eliminates the first two or three exons of c-abl and results in unregulated tyrosine kinase activity. The resultant fusion protein is either 210 kDa or 195 kDa. The latter version is more acutely transforming in experimental settings; it is also associated with acute lymphoblastic leukemia and with a worse prognosis in both disease settings. One of the ways in which the unregulated kinase activity may be manifest is through phosphorylation of SHC and/or GRB-2, adopter proteins necessary for coupling growth factor signals to ras.
c-jun This proto-oncogene encodes a ∼45 kDa transcription factor that is a member of AP1 family of transcriptional proteins. c-jun must form dimers to function and does so through the leucine zipper motif. Although c-jun-c-jun homodimers do form, they do so with low affinity and are not thought to be critical in gene transcription. Rather, a second partner, usually c-fos, generates the transcriptionally active heterodimer.
c-fos This ∼62 kDa leucine zipper protein cannot homodimerize but rather functions in heterodimeric complex with c-jun and other members of the AP1 family of transcription factors.
c-myc This proto-oncogene plays a critical role in hematopoietic cell proliferation. Like the leucine zipper protein, it too functions as a heterodimer. One of its partners is termed Max. The myc-related protein, Mad, also dimerizes with Max; the myc/Max complex stimulates proliferation, the Mad/Max complex inhibits myc-function. The importance of dysregulated myc function can be seen in Burkitt lymphoma in which a t(8;14) brings myc, on chromosome 8, into juxtaposition with the immunoglobulin enhancer on chromosome 14. Such upregulation of myc in a B lymphocyte setting results in a proliferative advantage and represents one important step in the genesis of this lymphoma. Myc has both leucine zipper and helix-loop-helix domains.
c-myb This gene encodes a transcription factor not belonging to any other class previously described and is expressed primarily in immature hematopoietic cells and declines as cells differentiate. Forced expression of c-myb tends to block hematopoietic differentiation. Clinically, high levels of myb are noted in acute leukemia, and such patients are less likely to enter remission or tend to have a short remission duration.
c-rel This gene belongs to the NF-κB family of transcription factors and can act to enhance or repress transcription from selected genes. This family of proteins includes p50 and its precursor p105, p65, p49 and its precursor p100, and Bcl-3, one of the IκB family.
IRF-1 (interferon regulatory factor-1) IRF-1 is a transcription factor that activates the expression of IFN α and β and maps to chromosome 5q31.1. As it is thought to act as a tumor suppressor gene, its role in the pathologic consequences of the 5q- syndrome is under active investigation.
IRF-2 (interferon regulatory factor-2) Interferon regulatory factor-2 is a gene which binds to a promoter element shared by IFN α and β and many IFN-inducible genes; unlike IRF-1, which stimulates such genes, IRF-2 represses transcription at the site. It is felt that the ratio of IRF-1 to IRF-2 might be a critical event in the regulation of cellular proliferation.
Rb The prototypical tumor suppressor gene Rb behaves in a genetically recessive fashion. Elimination or inactivation of both Rb gene copies is required for manifestation of the tumorgenic phenotype, first recognized in children with retinoblastoma. Such children inherit only a single functional copy; subsequent mutagenic inactivation of the remaining allele results in tumor susceptibility. Rb acts to sequester a group of transcription factors, termed E2F, which regulate genes critical for DNA synthesis. Alterations of Rb alleles are found in approximately 30% of human acute leukemias.
SCL This proto-oncogene, first identified in a stem cell leukemia at the site of t(1;14), is a member of the helix-loop-helix group of transcriptionally active proteins. The gene, also termed Tal 1, is expressed in erythroid and mast cell lineages but not in T cells. The association of t(1;14) with up to 25% of T cell ALL suggests that its ectopic expression is associated with transformation.
Bcl-1 This gene, located on chromosome 11 q13, was first identified at the site of translocation p(11;14)(q13;q32), has a strong association with central acinar/mantle cell lymphoma and functions in normal cells as the G1 cyclin termed CCND1 or cyclin D1. Normally, lymphocytes lack cyclin D1 expression; its aberrant expression resulting from chromosomal translocation of the Bcl-1 locus to an immunoglobulin locus is thought to be associated with aberrant proliferation.
Bcl-2 This gene product normally functions to suppress programmed cell death (apoptosis). Its overexpression is associated with the most common molecular abnormality in non-Hodgkin's lymphoma, t(14;18)(q32;q21), present in 80% of follicular small cleaved cell lymphoma. Presumably, suppression of apoptosis leads to extended cell survival, a characteristic of low-grade lymphomas.
Bcl-3 This gene is a member of the IκB family. Presently, it is unclear how this protein acts in tumorigenesis, but it is likely that its involvement in transcriptional processes is critical.
Bcl-6 A zinc finger transcription factor, expression of which is altered in approximately one-third of diffuse B large cell lymphomas as a consequence of 3q27 translocations. Its target genes are unknown.
RAR (retinoic acid receptor) The retinoic acid receptor is a member of the steroid hormone group of transcriptionally active proteins and contains a steroid hormone-binding domain, a zinc finger DNA-binding domain, and a transcriptional activation domain. RAR is located at the t(15;17) present in the majority of cases of acute promyelocytic leukemia. Its fusion partner in the translocation is termed pml. Normally, RAR forms heterodimers with members of the RXR family of transcription factors.
p53 Wild-type p53 is a sequence-specific DNA-binding nuclear protein that acts to induce gene expression. Overall, the program of p53-activated genes is associated with suppression of cell growth, consistent with our understanding of the mechanisms of anti-oncogenes. Mutations of p53 may not only inactivate its growth-suppression function, but can actually generate a genetically dominant, functional oncogene. Human tumors associated with p53 mutations include those of hematopoietic tissues (e.g., 20% of myelomas), bladder, liver, brain, breast, lung, and colon. It is likely the most frequently mutated gene in human cancer.
ras This gene encodes a critical signalling intermediate involved in the response to multiple growth factors. There are several related proteins (Ha-ras, Ki-ras, N-ras). N-ras and K-ras are mutated in many cancers, including 45% of myelomas and > 50% of CMML cases. Constitutive activation of ras can mimic chronic stimulation by the corresponding lineage-specific growth factor.
Hox 11 A homeobox containing transcription factor disrupted by translocation to the T cell receptor locus [t(10;14)] in 10% of cases of T cell ALL/lymphoblastic lymphoma. The Hox 11 gene is critical to the development of the spleen but its role in hematopoiesis is unclear.
Rhomb 2 Like Hox 11, Rhomb 2 is translocated in T cell ALL/lymphoma associated with t(11;14). Rhomb 1 may play a similar role in additional cases of T cell ALL. The Rhomb gene products are members of a family of transcription factors, but as Rhomb 2 and Rhomb 1 do not contain DNA-binding domains, they are thought to be involved in protein-protein interactions. Neither Rhomb 2 or 1 are normally expressed in T cells; transformation involving these genes, like SCL or Hox 11 is thought to be due to ectopic expression of the protein in T cells.
ALK (anaplastic lymphoma kinase) A large proportion of Ki-1 positive lymphomas are characterized by a t(2;5). The breakpoint involves nucleophosmin, a ubiquitously expressed gene, and ALK. The chimeric mRNA and protein are thought to be responsible for transformation. ALK is a member of the insulin receptor family of transmembrane receptor kinases, which is not normally expressed in hematopoietic tissues; the fusion gene is no longer membrane bound, which may underlie its pathogenesis.
Evi-1 A transcription factor whose rearrangement in t(3;21) is implicated as contributing to MDS. Overexpression of evi-1 blocks differentiation in response to hematopoietic growth factors.
ETO Located on chromosome 8, ETO is involved in t(8:21) of AML type M2. Based on the presence of two zinc-finger motifs. ETO possibly encodes a transcription factor, but its role in the pathogenesis of AML is unknown.
AML-1 Located on chromosome 21, AML-1 is the fusion partner of ETO in t(8:21). The gene is homologous to the runt gene of Drosophila and encodes a transcription factor. Normal hemopoietic targets include the CD13, GM-CSF, MPO, IL-3, and the T cell antigen receptor promoters. AML-1 binds as a heterodimer, partnered with CBFβ. It is unclear if its mechanism of action is to enhance aberrant transcription or to blunt transcription by acting in a dominant negative fashion.
CBFβ Located on chromosome 16, CBFβ is one of the fusion partners in the inv(16) associated with AML type M4Eo. As with AML-1, it is unclear whether the altered transcription factor enhances or blocks transcription.
MLL Located on chromosome 11, MLL (mixed lineage leukemia) is frequently altered in ALL, 1°AML, and especially in AML secondary to the use of topoisomerase II inhibitors. MLL is homologous to the trithorax gene of Drosophila and displays many features of a transcription factor and of a DNA methyl transferase.
Tel A helix-loop-helix transcription factor fused to the PDGFβ-receptor in CMML t(5:12) and to other genes in AML or MDS. Like most other translocation oncogenes, the mechanism of leukemogenesis is unknown. More recently, a Tel/AML1 fusion gene representing a t(12;21) has been found in a large number of cases of childhood ALL. As the translocation is not detected by routine cytogenetics, molecular analysis (FISH, etc.) is required to identify this favorable chromosomal rearrangement.
DEK Located on chromosome 6, DEK is involved in t(6:9) of AML. This translocation is usually seen in young patients and carries a poor prognosis. Its normal function is unknown, but DEK localizes to the nucleus.
CAN Located on chromosome 9, CAN is part of t(6:9). CAN forms part of the nuclear pore. As it has two different fusion partners but a consistent phenotype, CAN is likely the critical component of t(6:9).
Fas (CD95 or Apo-1) A transmembrane glycoprotein expressed on a wide variety of primitive and mature hematopoietic cells, which, upon binding to its natural ligand triggers programmed cell death.
NF-1 The gene responsible for neurofibromatosis. The normal protein functions to negatively regulate ras proteins, key intermediates in cytokine-induced cellular proliferation.
XI. Genetic Screening
X-linked methylation patterns Several loci present on the X chromosome become highly methylated when inactive but remain unmethylated on the active X chromosome (Lyon hypothesis). Should a polymorphic site for a methylation-sensitive restriction endonuclease exist at such an X-linked locus, one can distinguish between the active and inactive X chromosome by the pattern of restriction endonuclease digestion of that gene. However, in order to be widely useful for determining clonality of hematopoietic cells, the allelic frequency must be close to equality. Several X-linked genes meet these criteria and include phosphoglycerate kinase (PGK), hypoxanthine phosphoribosyltransferase (HPRT), the human androgen receptor gene (HUMARA), and the hyper-variable DXS255 locus. Both Southern blotting and PCR methods can be applied to this type of analysis.
RFLP (restriction fragment length polymorphism) If a mutation of one allele of a genetic locus either generates or destroys a restriction endonuclease site, the heterogeneity present within or very close to a gene of interest can be used to track which allele an individual has inherited from each parent. When genomic DNA is digested with a restriction enzyme that recognizes a polymorphic site and then hybridized with a probe specific for the gene of interest, the allelic pattern can be compared to that of a similar assessment of both parents. The presence of multiple family members allows a complete genetic pedigree to be constructed. For example, globin gene mutations such as sickle hemoglobin can be analyzed. The β6 mutation in hemoglobin, which results in Hgb S, destroys an Mst II site. Therefore, a larger than normal DNA fragment is generated by digestion of genomic DNA with Mst II, which can be easily detected by Southern blot hybridization. In this specific case, the Mst II polymorphism is absolutely specific for the mutant gene and family studies are not necessary. If the RFLP had not been specific for the mutation, but only existed close to the specific disease-producing mutation, then family studies would have been required to determine which pattern (presence or absence of restriction site) tracks with the mutant (disease) allele.
Allele-specific hybridization If the nucleotide basis for a specific genetic abnormality is known, oligonucleotides specific for wild type and for mutant sequence can be designed and used to probe Southern blots of an individual's genomic DNA. The pattern of hybridization thus gives specific information regarding which alleles are present. In a polymorphic disease such as β thalassemia (in which multiple mutations can give rise to the same disease phenotype), multiple probes might be required to detect all possible causes. In addition, new mutations causing the same disease would be missed. However, should a specific probe prove useful for one population group or be positive in one family member, that probe becomes very useful for the individual under study.
Reverse allele-specific hybridization This automated variant of allele-specific hybridization couples unlabeled synthetic oligonucleotides specific for a wild type or mutant sequence to a solid support that is then allowed to bind genomic sequences of the locus of interest, which have been amplified by PCR. The use of highly stringent conditions of hybridization allows differential binding of the amplified DNA to the wild type or mutant specific oligonucleotide and thereby allows genotypic determination of the individual.
Competitive oligonucleotide hybridization Mutant or wild type-specific oligonucleotide primers are used in a PCR reaction with genomic DNA. The primers and stringency of PCR are chosen so that single-based mismatches between genomic DNA and PCR primer fail to yield an amplified product. Thus, the PCR detection of a locus-specific product allows the genotyping of the individual.
Color complementation assay This method is an advancement over competitive oligonucleotide hybridization in that the wild type and mutation specific PCR primers are labeled with different color fluorescent tags, and both are used in a PCR reaction with genomic DNA. When highly stringent conditions are met, the fluorescent colors of the resultant PCR product indicate whether wild type, mutant, or both specific alleles were present in the original DNA sample.