1. Hospitalist, Leukemia Division, Section of Hematology-Oncology, University of Chicago Medical Center
2. Associate Professor of Medicine, University of Chicago Medical Center
Cytogenetically normal acute myeloid leukemia (CyN-AML) makes up 40 to 50 percent of cases of adult AML. The heterogeneous nature of CyN-AML makes prognosis uncertain, and despite the clinical heterogeneity, induction therapy remains largely uniform. Technological advances in DNA sequencing, however, are providing new insights into the molecular basis of CyN-AML, suggesting new diagnostic and prognostic categories and novel approaches to therapy.
In the case of CyN-AML, leukemogenesis is not driven by the consequences of a specific chromosomal rearrangement. Hence, investigators have focused on delineating molecular abnormalities that define disease subgroups. Mutations within the FLT3, NPM1, and CEBPA genes are now evaluated routinely at diagnosis, and the identification of one or more mutations in these genes gives prognostic information and aids in the selection of a consolidation strategy. In the case of FLT3 mutations, targeted therapies have been developed and are currently undergoing clinical trials.
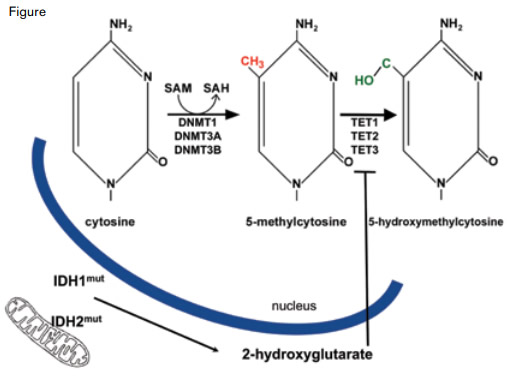
In addition to the three genes cited above, mutations in several genes encoding components of epigenetic pathways are common in CyN-AML: ASXL1 and MLL both encode modifiers of the histone proteins that compact DNA into chromatin, and DNMT3A, TET2, and IDH1/2 encode proteins that alter the balance of covalent cytosine modifications (Figure, page 5). Cytosines that are part of a 5'-CpG-3' dinucleotide are potential substrates for DNA methyltransferase (DNMT) enzymes, and ten eleven translocation (TET) proteins convert 5-methylcytosine to 5-hydroxymethylcytosine in an α-ketoglutarate (α-KG)-dependent reaction. Although the function of 5-methylcytosine as a transcriptional repressor is well established, the precise role of 5-hydroxymethylcytosine in facilitating transcription and/or as an intermediate in DNA demethylation is incompletely understood.
The TET gene family was originally identified when TET1 was discovered as a chromosomal fusion partner of MLL in AML, but the enzymatic function of the TET proteins was identified more recently. Although TET1 was discovered first, mutations in the TET2 gene are found more commonly in AML, ranging from 8 to 23 percent,1,2 and are thought to confer an unfavorable prognosis.3
Within the cell nucleus, DNA methyltransferase (DNMT) enzymes convert cytosine to 5-methylcytosine using S-adenosylmethionine (SAM) as the methyl donor. The TET proteins convert 5-methylcytosine to 5-hydroxymethylcytosine. Because the TET proteins are α-ketoglutarate-dependent enzymes, their activity is inhibited by 2-hydroxyglutarate, a metabolite produced in excess specifically in cells with mutated isocitrate dehydrogenase 1 or 2 (IDH1/2) proteins. While both IDH1 and IDH2 are found within the cytoplasm, IDH2 is also located in cellular mitochondria.
Within the cell nucleus, DNA methyltransferase (DNMT) enzymes convert cytosine to 5-methylcytosine using S-adenosylmethionine (SAM) as the methyl donor. The TET proteins convert 5-methylcytosine to 5-hydroxymethylcytosine. Because the TET proteins are α-ketoglutarate-dependent enzymes, their activity is inhibited by 2-hydroxyglutarate, a metabolite produced in excess specifically in cells with mutated isocitrate dehydrogenase 1 or 2 (IDH1/2) proteins. While both IDH1 and IDH2 are found within the cytoplasm, IDH2 is also located in cellular mitochondria.
The activity of TET2, an α-KG-dependent enzyme, is inhibited by 2-hydroxygutarate (2-HG), a metabolite produced in excess by mutated isocitrate dehydrogenase 1 and 2 (IDH1/2) enzymes. The IDH family of enzymes includes IDH1 and its mitochondrial isozymes, IDH2 and IDH3. The IDH enzymes catalyze the conversion of isocitrate to α-KG in the citric acid cycle, but mutations can confer a unique gain-of-function to the enzymes that results in excess production of 2-HG, a molecule usually found in trace amounts in cells. When levels are high, 2-HG diffuses throughout the cell (including the nucleus) and inhibits TET2 (Figure).
IDH1 and its mitochondrial variant, IDH2, first came to the attention of oncologists in 2006, when IDH1 mutations were found in colorectal adenocarcinoma. Most of the early attention, however, focused on gliomas and secondary glioblastomas as IDH1/2 mutations are found in > 70 percent of these tumors. IDH mutations have since been implicated in AML, particularly CyN-AML, with varying frequencies across case series – from 5.5 percent (based on 493 patients who had only IDH1 sequenced)4 to 33 percent (based on 358 patients who had both IDH1 and IDH2 sequenced).5
In the earlier glioma studies, oncogenic mutations were identified at the arginine 132 residue (R132) in IDH1, and its analog, arginine 172 (R172), in IDH2. Notably, these mutations were found to be mutually exclusive in gliomas. A third IDH2 mutation, arginine 140 (R140), was found to be AML/MDS-specific. Concurrent IDH1 and IDH2 mutations in AML have been reported to be either mutually exclusive6 or very rare,5,7 and concurrent TET2 and IDH mutations have not been identified.8 These observations lend credence to a model in which IDH mutations lead to low α-KG and high 2-HG levels in leukemic cells, thus inhibiting TET2. As expected, TET2 and IDH1/2 mutant AMLs have a similar hypermethylation phenotype as in neither case is 5-methylcytosine efficiently converted to 5-hydroxymethylcytosine.8
This model has diagnostic, prognostic, and therapeutic implications. Diagnostically, it may be possible in the future to measure 2-HG levels within leukemic cells as a means of diagnosing IDH1/2 mutations. Several studies have examined the impact of IDH1/2 mutations on prognosis. Although the conclusions are not unanimous, the largest dataset consisting of > 1,000 patients enrolled on the UK MRC AML 10 and AML 12 trials led to conclusions supported by some smaller studies — namely that patients with the IDH2-R140Q mutation had a more favorable prognosis, while the IDH2-R172 mutation appears to be neutral prognostically, and the IDH1-R132 mutations confers a worse prognosis.2,7,9,10 This series took the mutational status of FLT3, NPM1, and CEBPA into account in multivariant analysis, and future studies will also need to analyze a panel of gene mutations to clarify whether interactions involving these abnormalities affect prognosis.
Because the oncometabolite 2-HG is markedly elevated in IDH1/2-mutated cells, testing through non-invasive means (e.g., serum/urine levels) may be used in the future for monitoring of AML disease progression and/or relapse. Therapeutically, drugs that target IDH1/2-mutated AMLs are under development. Disappointingly, the first small trials using hypomethylating agents in TET2-mutated disease showed either no impact11 or worse survival.12 Further investigations of these and other mutations are ongoing and will likely affect clinical decisions in the near future. Unraveling of the complex mechanisms involved in epigenetic modification of DNA and chromatin remodeling has provided surprising new insights into the biology of neoplastic diseases. From this dynamic field of investigation, we can look forward to the discovery of other novel, unanticipated pathologic processes that will ultimately affect the diagnosis, prognosis, and management of hematologic malignancies.
References
Competing Interests
Drs. King and Godley indicated no relevant conflicts of interest.