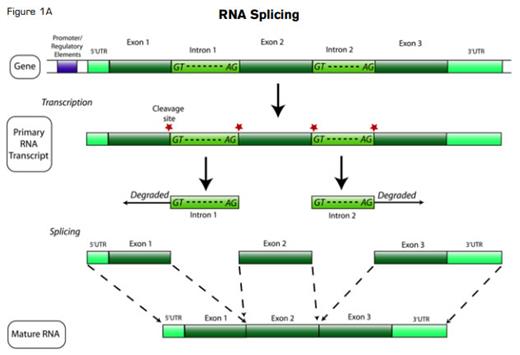
Of all the molecular processes that a biologist-detective might interrogate for the cellular crime of promoting malignant behavior, surely pre-mRNA splicing — i.e., knitting together coding information-bearing exons and discarding “useless” introns to produce a mature gene transcript, ready for translation into protein — must rank among the least likely suspects.
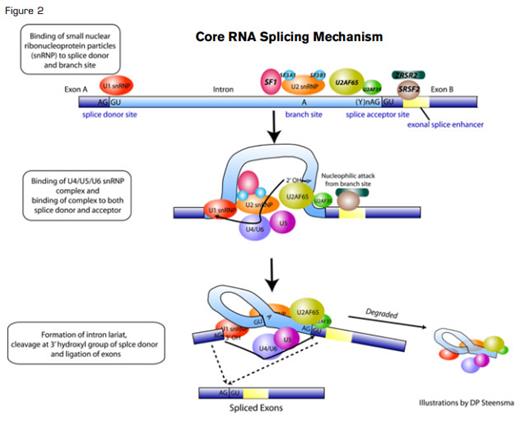
There has been, admittedly, circumstantial evidence pointing toward at least an accomplice rap for RNA splicing in cancer promotion: Aberrantly and alternatively spliced transcriptional isoforms are commonly found in the transcriptome of neoplastic cells, and transformed cells often selectively express specific transcripts (e.g., encoding more active kinase isozymes) that can confer a growth or survival advantage. But almost all metazoan genes are multi-exonic, and ~95 percent of eukaryotic multi-exonic genes are alternatively spliced using the same highly conserved splicing machinery (Figure 1A). So how could that basal splicing machinery itself possibly be deranged without causing catastrophic cellular consequences? The mRNA splicing process itself is so fundamental to normal cell behavior that any serious defect in the small nuclear ribonucleoproteins (snRNPs) or the >100 associated protein factors comprising the spliceosome that catalyzes splicing should be lethal.
Many investigators presumed that cancer-associated alternative splicing resulted from cis-acting DNA mutations in conserved intron/exon boundary recognition sites (Figure 1B), or another local phenomenon, such as a specific epigenetic pattern favoring one splicing isoform over another (since splicing is co-transcriptional). All previously known mutations in trans-acting splicing factors appeared to have only cell-specific effects, such as the germline pre-mRNA processing factor (PRPF) mutations underlying certain forms of autosomal dominant retinitis pigmentosa.
But now, new incontrovertible evidence has surfaced — the oncologic equivalent of a smoking gun or a bloody glove that fits. Several ongoing next-generation sequencing projects (ironically, whole-exome resequencing) have identified a high frequency of somatic mutations in core splicing components in myelodysplastic syndromes (MDS) and in secondary acute myeloid leukemia (AML, either post-MDS or therapy-related); the same mutations are also found at a low rate in numerous other neoplasms.1-3
At least eight different splicing components — SF3B1, SRSF2, U2AF35, ZRSR2, SF3A1, PRP40B, SF1, and U2AF65 — are somatically mutated in patients with MDS, always heterozygously and almost always mutually exclusively.2 Collectively, mutation in one of the components of the spliceosome is found in 45 to 85 percent of MDS and chronic myelomonocytic leukemia (CMML) cases. The most frequently abnormal gene, SF3B1, is mutated in 20 to 45 percent of MDS cases generally and, notably, in 65 to 85 percent of MDS cases associated with ring sideroblasts — a striking morphologic-genetic correlation.1,2 Acquired mutations in these same splicing factors are also found at least occasionally in a series of other neoplasms, including 9 to 15 percent of chronic lymphoid leukemia (CLL);4,5 3 to 9 percent of myeloproliferative neoplasms and de novo AML;1,2,6 and rare cases of breast, renal, and adenoid cystic carcinomas – and probably other tumor types as well.1
In MDS, several groups have reported that splicing mutations are associated with longer overall survival and leukemia-free survival,1,7,8 though a Mayo Clinic series suggests that the presence of a splicing mutation may not be independent of known MDS prognostic features such as morphology or the International Prognostic Scoring System (IPSS) score.9 In CLL, in contrast, SF3B1 mutations are associated with poorer prognosis that correlate with ATM mutations and deletion of chromosome 11q.4
Strangely, all eight of the known mutated splicing factors are involved in recognition of the canonical 3’ DNA splice element and its nearby polypyrimidine tract, while no mutations have yet been described in 5’ elements (e.g., U1snRNP components). Even more peculiarly, mutations in SF3B1 and mutations in DNMT3A, which encodes a DNA methyltransferase and is associated with poorer outcomes in MDS, appear to co-associate more frequently than would be expected by chance alone.7 It is not clear why this should be the case.
A host of new questions are raised by these findings.10 First, what are the molecular consequences of splicing machinery gain-of-function or loss-of-function mutations? Although there is considerable allelic heterogeneity, SF3B1 K700, and H662 represent high-frequency recurrent mutation sites. Other detected mutations are less deleterious than would be predicted if the mutations occurred randomly, and null mutations are rarely if ever observed.2 These observations, that mutations are not simple loss of function but instead lead to aberrant spliceosomal properties, suggest that perhaps neomorphic alleles result from the mutations in a manner analogous to the AML-associated IDH1 R132H mutation.
Second, why are mutations only found in 3’ splicing elements in MDS, CLL, and other diseases? Is there something special about 5’ elements; would changes there be lethal to the cell?
Third, a similar question arises as that raised by ribosomal defects in MDS (particularly in relationship to the 5q- syndrome) — does a splicing mutation result in a clonal advantage in the marrow milieu, and if so, how? Initial experimental results do not suggest that mutant spliceosome components confer a proliferative or survival advantage; transfection of mutant U2A35 into HeLa and TF 1 cell lines reduced the cells’ proliferative potential and increased apoptosis compared to cells transfected with wild-type U2A35, while mutant-transduced murine stem cells exhibited compromised reconstitution capacity in lethally irradiated mice.2 (SF3B1 cDNA is challenging to work with and results of transfections have not yet been reported.) Interactions with the polycomb repressor complex suggest a possible non-splicing role for SF3B1.
Fourth, why are splicing mutations so tightly associated with ring sideroblasts? Is there a gene already associated with sideroblastic anemia, such as ABCB7, SLC25A38, or ALAS2, mis-spliced in a consistent way? Transfection of mutant splicing factors into cells increased generation of unspliced RNA species with a premature stop codon and induced nonsense-mediated decay, but no specific altered transcript of interest was observed.2 Clearly, there is much work still to be done to characterize these mutations functionally.
Finally, is there any way disordered splicing could be a therapeutic target? There are several compounds that can alter splicing patterns, but it is too early to predict whether any of these reagents might have clinical utility.
The MDS splicing story rings a personal note. In 2007, puzzled by the frequency of expressed aberrant spliceoforms of genes such as CDC25C and ATRX without any correlating genomic changes in canonical splicing elements,11,12 I submitted an R01 grant application proposing to look for mutations in spliceosome components in MDS primary cells (proposal R01 CA136631-01). The suspicious biologist-detectives of the Cancer Molecular Pathobiology study section were appropriately skeptical of my application, and the proposal was triaged without a score. One reviewer commented that splicing was an implausible target in MDS – and besides, if disordered splicing were truly important, more well-established labs would surely already be working on the problem.
As a relatively callow young scientist, I lost confidence in my ideas, assumed the study section was correct and that I was barking up the wrong tree, shut down my lab, and moved to a clinical job at a new institution. Four years later, to see MDS-associated splicing mutations become Nature and New England Journal of Medicine papers and an ASH annual meeting plenary session makes me happy for friends and colleagues involved, and glad progress is being made, yet disappointed not to be part of the action – and kicking myself for giving up too soon. Erstwhile independent U.S. Presidential candidate H. Ross Perot once said, “Most people give up just when they’re about to achieve success. They quit on the one yard line. They give up at the last minute of the game, one foot from a winning touchdown.” The take-home message for young investigators: Don’t let that be you, too.
References
Competing Interests
Dr. Steensma indicated no relevant conflicts of interest.