Professor and Chair, Department of Medicine, University of Florida Health Science Center, Shands Hospitals
Dr. Robert Hromas is the Scientific Advisor for Northlake Biosciences, LLC.
Cancer is a disease of DNA repair. Since every cancer must have at least one mutation that produces the characteristic uncontrolled proliferation that defines neoplasia, at least one instance of abnormal DNA repair must have occurred in every cancer. Indeed, the genomes of cancers sequenced thus far have demonstrated multiple DNA mutations1 implying that abnormal DNA repair can be both the cause and the effect of neoplastic transformation.
For some cancers, the defect in DNA repair is obvious. For example, inherited mutations of the Ataxia Telangiectasia-Mutated (ATM) gene, the NBS1 gene, or Fanconi Anemia genes can lead to acute myeloid leukemia.2 Perhaps the best known example of a defect in DNA repair leading to cancer is BRCA1/2 mutations in breast, ovarian, and peritoneal malignancies.3 What is just now beginning to be understood is that all cancers, not just the familial malignancies, will have some aspect of DNA repair that is less functional than normal. However, discovering the specific defects in DNA repair for each tumor type, or even between individuals with the same tumor type, will take lengthy investigation.
The paradox that all cancer cells face is that to replicate they need DNA repair. Each replication fork moving along the DNA strand will use several DNA repair pathways just to maintain DNA synthesis. The replication fork can hit strand breaks, cross-links, or damaged nucleotides, all of which occur commonly in all types of cells, and it stalls at these blockades. To restart the replication fork, the cell must repair these blocking lesions. How can this occur if all cancers have some underlying abnormality in DNA repair?
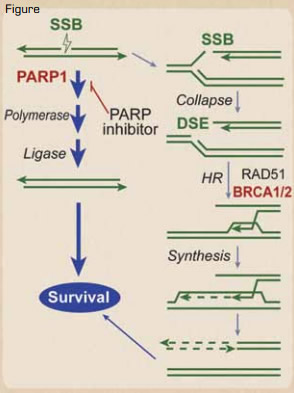
Synthetic Lethality in BRCA1/2 Defective Tumors. Naturally occurring single-strand breaks (SSBs) are primarily repaired by a PARP1-dependent pathway before a replication fork hits. However, if they are not repaired before the replication fork hits, the SSB is converted into a DSB. The DSB at the fork is usually repaired by a type of DSB repair termed homologous recombination (HR), which requires BRCA1/2, denoted in red. Thus, BRCA1/2 are needed to restart the collapsed replication forks caused by the DSB. In BRCA1 or 2 defective tumors, these SSBs must be repaired by the PARP1 pathway before the replication fork hits, as there is no mechanism of repairing the resultant DSB. Thus, PARP inhibitors result in synthetic lethality of the BRCA1 or 2 defective cancer cell, because the SSB cannot be repaired in front of the replication fork, and the resultant DSB caused by the progression of the fork through the SSB also cannot be repaired. Normal tissue, which does not have the bi-allelic defects in BRCA1 or 2, would not be affected.Reprinted from Blood. See reference number 4.
Synthetic Lethality in BRCA1/2 Defective Tumors. Naturally occurring single-strand breaks (SSBs) are primarily repaired by a PARP1-dependent pathway before a replication fork hits. However, if they are not repaired before the replication fork hits, the SSB is converted into a DSB. The DSB at the fork is usually repaired by a type of DSB repair termed homologous recombination (HR), which requires BRCA1/2, denoted in red. Thus, BRCA1/2 are needed to restart the collapsed replication forks caused by the DSB. In BRCA1 or 2 defective tumors, these SSBs must be repaired by the PARP1 pathway before the replication fork hits, as there is no mechanism of repairing the resultant DSB. Thus, PARP inhibitors result in synthetic lethality of the BRCA1 or 2 defective cancer cell, because the SSB cannot be repaired in front of the replication fork, and the resultant DSB caused by the progression of the fork through the SSB also cannot be repaired. Normal tissue, which does not have the bi-allelic defects in BRCA1 or 2, would not be affected.Reprinted from Blood. See reference number 4.
Typically, cancer cells resolve this paradox by overusing an alternative DNA repair pathway that can achieve the same end. In other words, in order to replicate, cancer cells become addicted to a defined DNA repair pathway other than the one that was defective in the origin of their transfomation.4 This means that cancer cells have a fundamental weakness that their normal counterparts do not have. Targeting the alternative pathway to which a given cancer is addicted can stop replication forks from progressing, without affecting the replication of normal cells. This novel and potentially revolutionary therapeutic insight is called “synthetic lethality.”4
The best example of this is in the hereditary breast and ovarian cancer syndromes mentioned earlier. These BRCA1/2-deficient tumors are defective in the repair of DNA double-strand breaks (DSBs). When a replication fork in one of these tumors hits a DNA single-strand break (SSB), it converts that into a DSB, but the replication apparatus cannot progress past that DSB. Since BRCA1/2 are both required for DSB repair, the tumor cells with those mutated repair components will rely heavily on, even be addicted to, repair of SSBs to prevent these DSBs from occurring. These cancer cells must repair the SSB before a replication fork hits it and converts it into a DSB. The DNA repair protein PARP1 is required for repair of SSBs, and there are small molecular inhibitors of PARP1 that will prevent repair of SSBs, but this is only a problem for the cells that are deficient in BRCA1/2 (Figure) — in this case, the tumor cells. Normal cells have the ability to repair the DSBs generated at the replication fork, because they have at least one normal allele of BRCA1/2. This has been proven clinically, where the PARP1 inhibitor olaparib improves the progression-free survival of familial breast cancer.5
Of course, not all breast or ovarian tumors have mutant BRCA1/2, but many more tumors behave as if they have such defects. For example, many ER-negative, PR-negative, and Her-2-negative (triple negative) breast cancers behave as if they were defective in BRCA1 or 2. In these patients, inhibiting PARP1 produced superior outcomes than not inhibiting PARP1.6 These patients may not have mutations in BRCA1 or 2, but they likely had deficiencies in other steps in this DSB repair pathway that generated the same characteristics. In other words, just because a given cancer is sporadic as opposed to familial does not mean it does not have a defect in DNA repair.
Tumor cells that are defective in DNA repair are clever, though. They can escape therapy based on synthetic lethality via the inherent instability of their own DNA. The best example of this is that some BRCA2-mutated breast cancers treated with PARP1 inhibitors generate a mutation leading to re-expression of a functional BRCA2 gene.7 These breast cancer cells with mutated BRCA2 actually deleted the mutated part of the gene under the selective pressure of PARP1 inhibitor therapy. This resulted in a smaller but functional BRCA2 protein, which could now repair the extra DSBs that occurred during replication in the presence of PARP1 inhibition.
Lest one think that targeting DNA repair for cancer therapy is solely the province of solid tumor oncology, there are several examples that indicate its applicability to hematologic neoplasia as well. Bortezomib synergizes with the alkylating agents in the treatment of myeloma by inhibiting DNA repair,8 and there is a subset of CLL with poor prognosis that has mutations in the ATM gene,9 which is also important in DSB repair. These CLL cells have been found to be amenable to therapy with PARP1 inhibitors. Finally, AML cells can be sensitive to PARP1 inhibition because of their intrinsic genomic instability.10
In the future, it may be that every individual’s cancer genome will be sequenced. This would better define which defective DNA repair pathways to target using the synthetic lethality approach. Once such defective pathways are defined for all tumor types, drug development can target the alternative, required pathway. In addition, this information can be used as a biomarker to predict response to these agents. Thus, targeting DNA repair to enhance cancer therapy is one of the most innovative advances in cancer drug development in the last decade and may become widely applied in many areas of oncology.