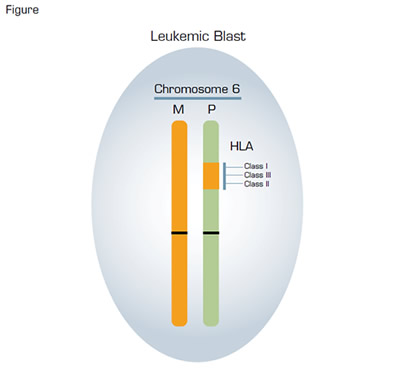
Acquired Uniparental Disomy in a Leukemic Blast in a Patient in Relapse Following Haploidentical Transplant. In the example illustrated, the original mismatched HLA alleles were located on the paternal (P) chromosome 6. During mitosis, a recombinant event occurred in one of the leukemic blasts in which a portion of the long arm of chromosome 6 containing the HLA loci was exchanged between a maternal and a paternal chromosome 6. Subsequently, a leukemic blast derived from this process contained the original maternal (M) chromosome 6 (orange) paired with the recombinant paternal chromosome 6 (green). That particular leukemic blast acquired a survival advantage in the setting of haplounidentical transplantation because it no longer expressed the mismatched paternal HLA proteins, thereby escaping immune surveillance by donor T cells that were haploidentical at the maternal HLA loci. Such recombinant events are not detected by standard karyotyping because there is no observable net loss or gain of genetic material. However, this process can be identified by using single nucleotide polymorphism-array analysis and is called copy number neutral loss of heterozygosity.
Acquired Uniparental Disomy in a Leukemic Blast in a Patient in Relapse Following Haploidentical Transplant. In the example illustrated, the original mismatched HLA alleles were located on the paternal (P) chromosome 6. During mitosis, a recombinant event occurred in one of the leukemic blasts in which a portion of the long arm of chromosome 6 containing the HLA loci was exchanged between a maternal and a paternal chromosome 6. Subsequently, a leukemic blast derived from this process contained the original maternal (M) chromosome 6 (orange) paired with the recombinant paternal chromosome 6 (green). That particular leukemic blast acquired a survival advantage in the setting of haplounidentical transplantation because it no longer expressed the mismatched paternal HLA proteins, thereby escaping immune surveillance by donor T cells that were haploidentical at the maternal HLA loci. Such recombinant events are not detected by standard karyotyping because there is no observable net loss or gain of genetic material. However, this process can be identified by using single nucleotide polymorphism-array analysis and is called copy number neutral loss of heterozygosity.
Haploidentical transplantation could just as easily be called haplounidentical transplantation, because recipient and donor are matched at HLA loci on one chromosome 6 (haploidentical) but not on the other (haplounidentical) (Figure). For treatment of patients with hematologic malignancy, transplanted cells serve two purposes. First, transplanted hematopoietic stem cells rescue the recipient from the myeloablative injury induced by lethal doses of chemotherapy and/or radiotherapy that comprise the preparative regimen. Second, transplanted lymphocytes are the source of the graft-versus-tumor effect that is essential for eradication of residual chemotherapy/radiation-resistant malignant cells. Therefore, the haplounidentical part of the equation is equally important therapeutically as the haploidentical part, as it is the mismatched HLA proteins that target the leukemic blasts for destruction by donor T cells. Haploidentical transplantation has the advantage over matched-sibling donor transplantation of higher probability of donor availability, as both parents and half of all siblings will be “matches.” On the downside, however, is the greater risk of high-grade graft-versus-host disease (GVHD) because all host tissue will be haplounidentical in relation to the donor lymphocytes. Despite the seemingly insurmountable task of transplanting across HLA barriers, haploidentical transplantation has become an important part of the hematologist’s armamentarium for treatment of high-risk and relapsed acute leukemia and relapsed lymphoma, and insightful translational research has led to clever new, more effective strategies for ameliorating GVHD while allowing for robust immune reconstitution.
In Brief
Regrettably, however, not all is good on the haploidentical transplantation front, as recently, investigators from Milan led by Katharina Fleischhauer and Fabio Ciceri reported an unexpected victory of sorts for the bad guys. Those investigators longitudinally evaluated donor-host hematopoietic chimerism in 43 patients who had undergone a haploidentical transplant. They were looking for the reappearance of recipient hematopoiesis that often heralds disease relapse, and they used both microsatellite markers and HLA typing for their analysis of chimerism. Seventeen patients (14 of whom received transplants when they had persistent disease) had a leukemic relapse, and in all cases, the leukemic blasts were found to be of recipient origin based on microsatellite analysis. But, surprisingly, no HLA proteins unique to the recipient were detected on the bone marrow cells of five of the relapsed patients. In other words, the haplounidentical antigens were no longer detectable on the leukemic blasts of those five patients despite the fact that microsatellite markers had shown that the cells were of host origin. The Milan group investigated the molecular basis of this phenomenon using single nucleotide polymorphism-array analysis and found that the leukemic blasts had acquired their homozygous HLA phenotype through the process of uniparental disomy (Figure). This remarkable outcome is a vivid example of Darwinian evolution in the microcosm of the bone marrow. In this case, donor T cells exerted a powerful selection pressure on the host bone marrow cells, destroying the cells based on recognition of the haplounidentical HLA proteins. Only the leukemic blasts that had eliminated the mismatched HLA loci through uniparental disomy escaped immune surveillance (Figure). Armed with their understanding of the molecular basis of relapse in these patients, the investigators subjected two of the patients to a subsequent transplant using cells from a different donor who was mismatched for the HLA haplotype retained by the leukemic cells. Remarkably, one of the two patients was alive and in complete remission more than 16 months after the second transplant. Together, these observations demonstrate a novel mechanism for development of treatment resistance in patients undergoing haploidentical transplant. That the process occurred in 5 out of 17 (29 percent) of the relapsed patients indicates that this mechanism is a relatively common cause of treatment failure. Importantly, understanding the basis of the resistance allows for a tailored approach to treatment of relapse and hope for long-term survival for affected patients — a victory for the good guys.
"Be Good and You Will Be Lonesome" - Quote by Mark Twain. 2010 marks the centennial of Twain’s death (November 30, 1835 - April 21, 1910).
Competing Interests
Dr. Parker indicated no relevant conflicts of interest.