Gondek LP, Tiu R, Haddad AS, et al. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in MDS. Leukemia. 2007;21:2058-61.
With the completion of the Human Genome Project and its sister HapMap Project that mapped single nucleotide polymorphisms (SNPs), the last few years have seen a revolution in how information about DNA sequences and genotypic variation are utilized in research. Soon, all these will be transferred to the clinical arena. The development of genotyping platforms, such as SNP chips, has allowed simultaneous assessment of hundreds of thousands of SNPs, as well as copy-number variations and the determination of the nature of loss-of-heterozygosity including uniparental disomy (UPD) across the whole human genome. High-resolution, genome-wide SNP chips, following the example of other high-tech devices, are including an ever-increasing number of SNPs at a rapidly declining cost. They are being increasingly used in association studies of inherited predispositions for both rare and common human diseases.1 However, three recent studies also highlight their use for the identification of somatic point mutations and cryptic chromosomal lesions in blood disorders when applied to large cohorts of patient samples.
In an important study, Mullighan and colleagues describe an unprecedented genome-wide SNP analysis of 242 pediatric patients with acute lymphoblastic leukemia (ALL) using high-resolution SNP chips. This newer generation of chips features detections of more than 350,000 loci and permits analysis of copy-number changes at an average resolution of <5kb across the entire human genome. The study identified 54 recurrent somatic regions of deletion, of which 24 contained only a single gene. Further analysis of selected genes from these regions revealed that 40 percent of patients had deletions or mutations in genes that control B lymphocyte development and differentiation, with the PAX5 gene being the most frequent target of somatic mutation (in 31.7 percent of patients). The study also found deletions in additional genes important in B-cell differentiation including EBF1 and IKZF1 (IKAROS), suggesting a contribution of these genes to the pathophysiology of B-progenitor ALL.
Though examining a different clonal hematologic malignancy, myelodysplastic syndrome (MDS), two other papers published this year explored the usefulness of SNP chips in studies of acquired genetic imbalances. In MDS, a considerable percentage of patients do not exhibit cytogenetic abnormalities using conventional metaphase chromosome analysis. High-resolution SNP chips, however, offer improved levels of genomic resolution, facilitating detection of both cryptic defects as well as their copy number. In independent studies, Mohamedali et al. and Gondek et al. carried out genome-wide SNP analysis in 119 and 66 cases of MDS, respectively. Surprisingly, both groups identify a high frequency of segmental loss-of-heterozygosity due to UPD, which is not detectable by traditional cytogenetic analysis. UPD results from duplication of one of the parental alleles during mitotic recombination but is not detectable by cytogenetic analysis. UPD was previously found to be common in polycythemia vera2 and other myeloproliferative disorders, but had not been appreciated in other hematologic malignancies. These three studies confirm the greater power of SNP chips over the cumbersome metaphase-dependent cytogenetics, bypassing the need for laborious low-resolution and costly metaphase examination. It remains to be established whether SNP chips could avoid the need for bone marrow tissue since easily accessible clonal circulating granulocytes may provide the same information as analysis of hematopoietic progenitors.
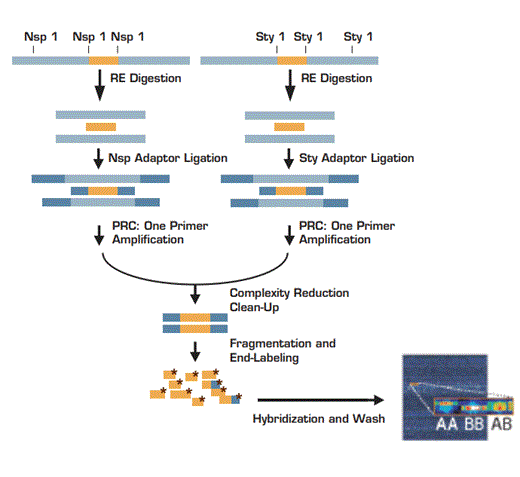
Step 1: Individual digestion of genomic DNA (500ng) with Nsp I and Sty I restriction enzymes (RE). Step 2: Ligation of digested products to corresponding adaptors. Step 3: Preferential amplification of adaptor-ligated RE digested DNA fragments with generic primer to generate products of sizes from 200 to 1100 bp. Step 4: Combination of PCR products from each RE digest. Step 5: Purification of products using polystyrene beads. Step 5: Fragmentation and labeling of the amplified DNA. Step 6: Hybridization of labeled fragments to SNP chip.
Step 1: Individual digestion of genomic DNA (500ng) with Nsp I and Sty I restriction enzymes (RE). Step 2: Ligation of digested products to corresponding adaptors. Step 3: Preferential amplification of adaptor-ligated RE digested DNA fragments with generic primer to generate products of sizes from 200 to 1100 bp. Step 4: Combination of PCR products from each RE digest. Step 5: Purification of products using polystyrene beads. Step 5: Fragmentation and labeling of the amplified DNA. Step 6: Hybridization of labeled fragments to SNP chip.
In Brief
Taken together, these three pioneering manuscripts show the amazing ability of SNP chips to detect somatic mutations associated with acquired clonal hematologic disorders and demonstrate UPD as one of the most common genetic mechanisms found. These elegant studies highlight the increasing speed, specificity, and power of genome-wide approaches in the systematic search for pathogenic lesions. As the cost of SNP chips has recently dropped to within reach of most laboratories ($250 per sample), and as genome-wide chips are being produced with even higher densities (the current-generation chip contains 1 million SNPs), the way we investigate, diagnose, and stratify therapy for both germline and somatic mutations will likely be transformed. With these rapidly evolving technologies, the possibility of having the entire 3-billion-base-pairs human genome sequence hybridized to a single chip could actually come a lot sooner than we thought.
References
Competing Interests
Drs. Liu and Prchal indicated no relevant conflicts of interest.