The identification of somatic and germline genetic lesions of hematologic malignancies has contributed to our understanding of the biology of cancer. Further elucidation of the molecular mechanisms in cancer should eventually provide dramatic clinical benefits, just as the understanding of the molecular impact of the BCR/ABL translocation has fundamentally changed the prognosis and therapy of chronic myelogenous leukemia.
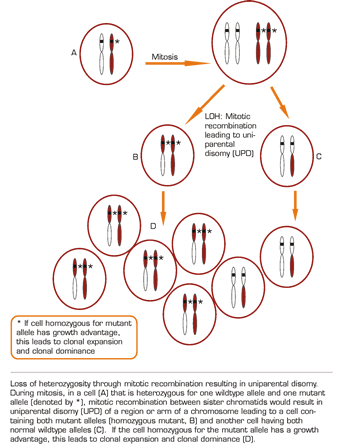
Tumor suppressor genes limit unregulated cell proliferation and maintain normal differentiation. Classically, they act in a dominant fashion such that a single normal allele is enough to avoid malignant transformation. Heterozygosity is having two dissimilar alleles at a locus. If this is at a tumor suppressor locus and one of these alleles is normal and one mutant, it will still result in normal differentiation and maturation. Loss of heterozygosity (LOH) is the reduction from two dissimilar alleles to, effectively, only one. LOH through loss of DNA occurs through deletions or partial or complete chromosome loss resulting in hemizygosity, or only a single copy of the locus. Conversely, LOH without loss of DNA can be a result of mitotic recombination or gene conversion resulting in homozygosity, or two copies of the same allele. Functional LOH represents expression of only a single allele with no change in the DNA. This is through epigenetic mechanisms such as DNA methylation, histone acetylation, or changes in subnuclear localization1 , all of which can affect the transcriptional activity of the affected gene(s). Disruption of the wildtype allele by genetic or epigenetic changes in the regulatory or coding regions may expose either loss- or gain-of-function in the other allele. Conventional cytogenetics (karyotyping), comparative genomic hybridization, and fluorescence in situ hybridization (FISH) can detect more substantial chromosome architectural abnormalities but cannot detect more subtle genetic changes, since these techniques do not have sufficiently high resolution or cannot detect LOH without loss of genetic material (epigenetic changes).
Recently, another mechanism by which LOH can occur has been identified — uniparental disomy (UPD) created by mitotic recombination. In this case, the consequences of LOH are fundamentally different from those described above in which loss of gene function is the pivotal event. In UPD, the mitotic recombination of a heterozygous cell with one mutant allele results in daughter cells that are homozygous for the mutant allele or contain two copies of the wildtype alleles (Figure). Thus, UPD is a condition wherein both homologues at a particular chromosomal region, or alleles at a particular locus, are derived from the same parent. If the cell homozygous for the mutant allele has a growth advantage, this leads to clonal expansion and clonal dominance. Some types of LOH can be identified by comparing normal and abnormal tissues from an individual with cancer and demonstrating the presence of heterozygosity at a particular genetic locus in normal tissue and the LOH in abnormal tissue. This is often done using polymorphic markers such as microsatellites and single nucleotide polymorphisms (SNPs); however, these DNA-based studies will not identify epigenetic LOH.
Using genome-wide microsatellite markers, our laboratory2 identified three genomic regions that had LOH in patients with polycythemia vera (PV). The most frequently observed segment of LOH (33 percent of PV patients) covered a region of about 40 megabases on the short arm of chromosome 9 (9pLOH). Cytogenetic analysis in these patients did not detect any deletion, while more detailed laboratory studies confirmed the presence of two copies of the same alleles in the 9pLOH region, suggesting that the LOH arose through mitotic recombination resulting in UPD. The JAK2 gene, found within this 9p region, plays a crucial role in erythropoietin signaling. However, our laboratory missed the possible causative role of this gene in genesis of PV because we concentrated only on the tyrosine kinase domain of this gene2,3 . This gene has since been found by others to be mutated in patients with PV, and G1849T resulting in a valine to phenylalanine substitution at position 617 (V617F) of the Jak2 protein was identified4 . A majority of patients with idiopathic myelofibrosis have 9pUPD and, like patients with PV, had at least a proportion of hematopoietic progenitors homozygous for the JAK2 V617F mutation. The JAK2 V617F mutation can also be demonstrated in approximately half of patients with essential thrombocytosis (ET)5,6,7 . In contrast to patients with PV and idiopathic myelofibrosis, however, evidence of JAK2 V617F homozygosity is almost never observed in patients with ET. This observation suggests that the phenotype of the myeloproliferative disorders is determined, at least in part, by gene dosage and raises the question of why UPD does not occur in patients with ET8,9,10 .
More recently, array-based genome-wide SNPs have been used to search for recurrent regions of UPD in hematologic malignancies. Analysis of LOH profiles in two series of adult patients with acute myelogenous leukemias (AML) having normal karyotype showed that about 10-20 percent had large regions of homozygosity11,12 . These regions of homozygosity due to somatically acquired UPD were observed in leukemic cells but could not be detected after a remission was induced. Acquired UPD as a recurrent genetic mechanism for LOH has also been shown to be prevalent in childhood leukemia, multiple myeloma, malignant lymphoma, and juvenile myelomonocytic leukemia13,14,15,16 . However, it is certainly not limited to hematologic malignancies. It has also been described in cancers of the breast, colon, lung, liver, gallbladder, and skin17,18,19,20,21 . These studies suggest that acquired UPD may be the primary common genetic mechanism by which LOH occurs in many malignant disorders. A potential consequence of UPD is that there may be the lack of active copies of a gene if the region of LOH contains a gene that undergoes genetic imprinting. Genetic imprinting of genes is established by selective inactivation of either the maternal or paternal allele. If a gene is imprinted, only one copy of the gene (depending on the parent of origin) is actively expressed. Preferential loss of the maternal 9p allele has been reported in childhood acute lymphoblastic leukemia22 . However, no evidence for parental bias in UPD was found in adult patients with AML11 , suggesting that these regions of LOH did not contain imprinted genes.
In spite of the heterogeneity of acquired genetic lesions within and between each type of cancer, 9pLOH occurs frequently in human cancer cell lines and some hematologic malignancies23 . It remains to be established whether this high frequency of 9pLOH (and the acquired UPD in cancers) occurs in a non-random fashion. There is growing evidence that the evolutionary conserved JAK/STAT signaling pathway may be responsible for 9pLOH UPD. A recent paper described a gain-of-function mutation of the JAK gene in Drosophila, suggesting that a mutant JAK protein appears to be involved in both cell proliferation and the global reversal of heterochromatic gene silencing24 , leading to carcinogenesis. It remains to be established whether gain-of-function mutations in the JAK gene may increase mitotic recombination — perhaps by increasing the probability of UPD in the 9p chromosome in myeloproliferative disorders — after a somatic V617F mutation has occurred25 .