The implications of identifying clonal hematopoiesis of indeterminate potential (CHIP) or clonal cytopenia of undetermined significance (CCUS) in an individual are not entirely clear. Both CHIP and CCUS fall on a spectrum with myelodysplastic syndromes/neoplasm and carry risk of progression. CHIP refers to the detection of somatic mutations in driver genes at a variant allele frequency (VAF) of 2% or greater in the absence of cytopenias, while in CCUS the mutations occur in the presence of an unexplained cytopenia. Both CHIP and CCUS lack defining features of myeloid neoplasm, including dysplasia below the threshold of 10%.1 Micro-clonal hematopoiesis (micro-CH) is a term applied to cases that otherwise fit CHIP but have a VAF below the threshold, i.e., between 0.5% and 2%.
While CHIP/CCUS is rare in individuals younger than 40, the incidence increases with age, and a substantial proportion of elderly individuals are affected.2 Patients with CHIP/CCUS uncommonly develop a myeloid neoplasm. The risk of progression is affected by the specific nature of the mutation(s) and size of the clone.3 Two scores, both published in 2023 and developed from UK Biobank participant data, attempt to model the risk of progressing from CHIP/CCUS to myeloid neoplasm. The Clonal Hematopoiesis Risk Score (CHRS) relies on the variables of mutation number (1 or ≥2), VAF (<0.2 or ≥0.2), red cell distribution width (<15 or ≥15), mean corpuscular volume (MCV, <100 or ≥100), cytopenia (CHIP vs. CCUS), age (<65 years vs. ≥65 years), and the presence or absence of a single DNMT3A mutation or high-risk mutation.4 The myeloid neoplasm prediction (“MN-predict”) score incorporates the parameters of gender, age, largest VAF, hemoglobin, MCV, and platelet count, in addition to the presence/absence of specific gene mutations.5 Both models are based on single-time point, cross-sectional clonal hematopoiesis (CH) measurement determinations. Longitudinal cohort studies, with DNA sequencing at multiple time points, have demonstrated variability in clonal expansion rate.6
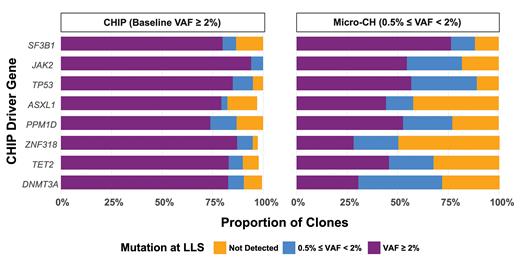
Yash Pershad and colleagues studied 6,986 participants in the Women’s Health Initiative Long Life Study who were sequenced at baseline (baseline median age ± interquartile range, 64 ± 9 years) and at an average of 15.8 years follow-up (range, 14-19 years). CH was identified by single-molecule molecular inversion probe sequencing assay in ASXL1, CBL, DNMT3A, GNB1, PPM1D, SF3B1, TET2, TP53, U2AF1, ZBTB33, ZNF318, SRSF2, IDH1, IDH2, and JAK2. There were 3,685 mutations with VAF of 0.5% or greater detected at baseline; of these, 23.6% were not detected at follow-up, 26.4% were micro-CH at followup, and 50.2% were CHIP at follow-up. Figure 1 presents the follow-up VAF percentage as it relates to baseline VAF and driver gene. Growth rate was estimated for mutations with baseline and follow-up VAF of 0.5% or greater using an exponential growth model (participants with more than one mutation were excluded, as this precludes use of the growth model).
Comparison of baseline variant allele frequency (VAF) to follow-up VAF by gene
Abbreviation: CHIP, clonal hematopoiesis of indeterminate potential; LLS, Women’s Health Initiative Long Life Study; micro-CH, micro-clonal hematopoiesis. The median ± interquartile range between baseline and follow-up was 15.8 ± 1.7 years.
Comparison of baseline variant allele frequency (VAF) to follow-up VAF by gene
Abbreviation: CHIP, clonal hematopoiesis of indeterminate potential; LLS, Women’s Health Initiative Long Life Study; micro-CH, micro-clonal hematopoiesis. The median ± interquartile range between baseline and follow-up was 15.8 ± 1.7 years.
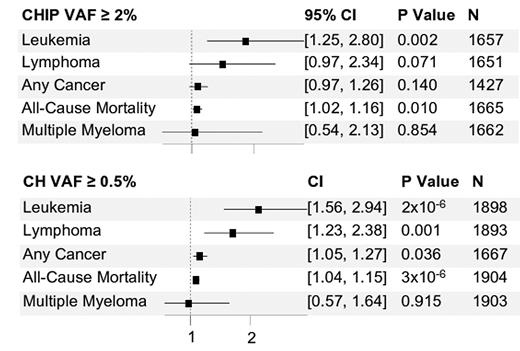
Data for the 1,906 women with baseline and follow-up CH revealed the fastest annual growth rates for JAK2 and SF3B1 (16.2% and 14.4%, respectively) and the slowest rates for DNMT3A and ZNF318 (6.9% and 8.3%, respectively). CH mutations of 0.5% or less grew more slowly than those of 2% or greater. The growth rate was independent of the baseline participant characteristics studied (e.g., age, smoking status, blood pressure, and body mass index, among others). The clonal growth rate in individuals with CHIP/micro-CH at baseline predicted cytopenia at follow-up, with faster growth rate associated with increased risk of cytopenia, adjusted for age and race. In time-to-event analysis, for both the cohort with VAF of 2% or greater and the cohort with VAF of 0.5% or greater at baseline, each 10% per year increase in growth rate was significantly associated with increased risk of leukemia and all-cause mortality (Figure 2). Further analysis tested whether the association between CHRS (calculated for follow-up point in time) and clinical outcomes was mediated by CHIP growth rate. There were significant mediation effects for leukemia (34.4% of the total effect of CHRS was mediated by growth rate) and a trend observed for all-cause mortality.
Hazard ratio per increase in growth rate by 10% per year
Abbreviations: CH, clonal hematopoiesis; CHIP, clonal hematopoiesis of indeterminate potential; VAF, variant allele frequency. For both the cohort with VAF of 2% or greater and the cohort with VAF of 0.5% or greater at baseline, each 10% per year increase in growth rate was significantly associated with increased risk of leukemia and all-cause mortality.
Hazard ratio per increase in growth rate by 10% per year
Abbreviations: CH, clonal hematopoiesis; CHIP, clonal hematopoiesis of indeterminate potential; VAF, variant allele frequency. For both the cohort with VAF of 2% or greater and the cohort with VAF of 0.5% or greater at baseline, each 10% per year increase in growth rate was significantly associated with increased risk of leukemia and all-cause mortality.
The authors also explored the association between leukocyte telomere length (LTL) and CHIP incidence/growth rate. Among individuals without CHIP at baseline, shorter baseline LTL was associated with increased risk of incident PPM1D CHIP; conversely, longer baseline LTL was associated with increased risk of incident DNMT3A CHIP. Baseline LTL did not associate with CHIP clone growth rate. However, for both the cohort with VAF of 2% or greater and the cohort with VAF of 0.5% or greater at baseline, the TET2 clone growth rate was associated with greater LTL shortening.
In Brief
The use of a sensitive assay to detect CH mutations at two time points in the same patient cohort provides information important to our understanding of CH progression. The study authors’ findings suggest a role for telomere biology in CHIP prevalence and expansion. Longitudinal data on micro-CH (VAF range, ≥0.5% to ≤2%) demonstrated similar growth rates and clone progression as seen in previous longitudinal studies, reinforcing the importance of clone size in addition to gene identity on progression risk. Future risk prediction scores may incorporate expansion rate into risk assessment for CHIP/CCUS.
Disclosure Statement
Dr. Courville indicated no relevant conflicts of interest.