I recently had a (very polite) argument with a hematologist. I reviewed a bone marrow biopsy performed on a 66-year-old man with mild thrombocytopenia and monocytosis, which did not show dysplasia or morphologic features of chronic myelomonocytic leukemia (CMML). However, next-generation sequencing (NGS) of his blood sample revealed pathogenic mutations in SRSF2, TET2 (two mutations), and RUNX1. The hematologist felt that the patient likely had CMML, but I was reluctant to make a diagnosis of myeloid malignancy. When I suggested that, clinically, this distinction should not matter because the patient’s mild cytopenias and lack of symptoms would not mandate therapy, the hematologist countered that they would appreciate more clarity to counsel the patient, guide follow up, and plan possible future care.
Identifying abnormal morphology is integral to a diagnosis of myelodysplastic syndrome (MDS) or CMML. Falling short of diagnostic morphology, individuals with cytopenia and clonal abnormalities detected via NGS are designated as having “clonal cytopenia of undetermined significance” (CCUS) — a nebulous category that confers an increased risk of subsequent myeloid neoplasia and shorter overall survival.1, 2 Blood NGS is increasingly being used in the diagnostic workup for unexplained cytopenia to screen for patients with a higher pre-test probability of myeloid malignancy on bone marrow biopsy; hence, it is expected that the number of patients being assigned a diagnosis of CCUS will increase.3, 4 NGS of solid tumors or “liquid biopsies” may also reveal CCUS (or, lacking cytopenia, clonal hematopoiesis of indeterminate potential [CHIP]) by identifying mutations in peripheral blood leukocytes.5 Thus, pathologists and hematologists alike are increasingly faced with clonal myeloid proliferations and the need to confront their border with hematologic malignancy. Yet, for individuals found to have CHIP or CCUS, the implications of this diagnosis on their future health trajectory remain uncertain.
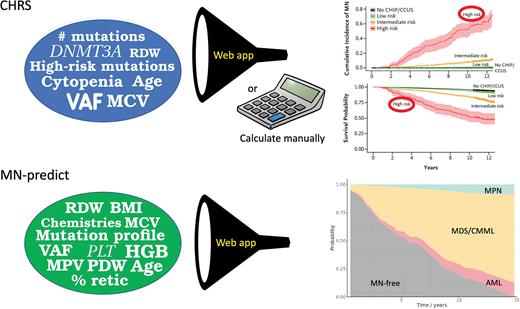
In their recent studies, Lachelle Weeks, MD, PhD, and colleagues and Muxin Gu, PhD, and colleagues examined a large number of samples from the UK Biobank (UKB), associating whole-exome sequencing genetic data as well as laboratory data at enrollment with the subsequent risk of developing a myeloid neoplasm (MN). Dr. Weeks and colleagues used separate subsets of data from 438,890 UKB participants to derive and validate their model. Among the 11,337 individuals found to have CHIP/CCUS, 2.37% developed an MN within a median follow up of 11.7 years. Having two or more mutations, a mutation variant-allele fraction (VAF) of 20% or more, the presence of any cytopenia, increased mean corpuscular volume (MCV) or red cell distribution width (RDW), and older age were all associated with an increased risk of MN. The model also incorporated the specific genes affected, with a single DNMT3A mutation being favorable and any mutations in SRSF2, SF3B1, ZRSR2, IDH1/2, FLT3, TP53, or RUNX1 being unfavorable. Their final model, the Clonal Hematopoiesis Risk Score (CHRS), includes three risk strata, with 10-year survival rates ranging from 93.7% for patients with low-risk disease to 51.2% for those with high-risk disease, as well as 10-year MN incidences ranging from 0.7% to 52.2%, respectively.
Dr. Gu and colleagues analyzed sequencing data from 454,340 UKB participants. In contrast to the approach used by Dr. Weeks and colleagues, their approach relied on separate models to predict the specific risk of progression to MDS or CMML (MDS/CMML), a myeloproliferative neoplasm (MPN), or acute myeloid leukemia (AML), since certain mutations and blood count abnormalities are associated with evolution to different subtypes of MN. The model developed by Dr. Gu’s team also incorporated additional variables such as platelet volume and distribution width, as well as blood chemistries (e.g., cholesterol and creatinine). Their final “MN-predict” model shows the risk of progressing from CHIP/CCUS to MDS/CMML, AML, or MPN over time. For individual patients, the CHRS can be calculated manually by adding points conferred by each variable or using an online app (www.chrsapp.com), while the MN-predict score is generated using an online app at bioinf.stemcells.cam.ac.uk/shiny/vassiliou/MN_predict.
These two studies have advanced our ability to predict the risk conferred by CHIP and CCUS, thus refining the implications of clonal hematopoiesis for an individual patient. Although these studies were population-based, Dr. Weeks and her team externally validated the CHRS in a series of patients with CHIP/CCUS referred to hematology clinics at two large medical centers. Not unexpectedly, high-risk cases were much more prevalent in these clinical cohorts (29.6% to 34.3% versus 1.1% in the UKB cohort); nevertheless, the CHRS model effectively predicted progressively higher incidences of MN among individuals with low-, intermediate-, and high-risk disease in both clinically referred cohorts.
These studies are expected to significantly influence the diagnosis and treatment of patients with hematologic disorders in several ways. (1) For individual patients and their treating physicians, the CHRS and MN-predict score refine the implications of a CHIP/CCUS diagnosis, facilitating more informed clinical follow up. (2) The patterns of evolution from CHIP/CCUS observed by Dr. Gu and colleagues shed light on predetermined clonal trajectories that lead to the major MN subtypes. (3) The strikingly high rate of “progression” from high-risk CHIP/CCUS to MN begs the question as to whether high-risk CCUS is truly different from chronic MN (e.g., MDS and CMML).6 On this note, evaluation for dysplasia is inherently subjective, and the dichotomous “dysplasia present versus absent” distinction between MDS and CCUS may not be optimal for guiding patient management.
Getting back to my argument with the hematologist, I attempted to prove my point by entering the patient’s clinical and genetic information into the CHRS and MN-predict models. To my surprise, despite the patient’s mild cytopenia and normal bone marrow morphology, the MN-predict tool indicated a 64% risk of developing MDS or CMML within 10 years, while the CHRS tool indicated a 10-year overall survival probability of only 51% (Figure). These results likely reflected the presence of high-risk mutations, high mutation VAFs, and abnormal clinical parameters such as an elevated MCV. Knowing that this patient has a high likelihood of developing MDS or CMML provided valuable information regarding clinical follow up and informed the hematologist’s conversations with the patient. These population-based models may be less applicable in some settings, such as when patients have been exposed to cytotoxic chemotherapy.7 Moreover, at present, MN-predict and CHRS do not have direct implications for treatment. Nevertheless, in the future, early identification of high-risk pre-MN disease states may allow the application of therapies that prevent clonal evolution and progression, potentially suppressing the risk of developing an MN and the need for more intensive therapies.8
The CHRS model uses input variables (blue circle) to calculate the risk of developing any myeloid malignancy and predict overall survival. Inputting the variables from my patient indicated a high-risk assignment. The MN-predict model uses similar input variables as the CHRS model, although several additional clinical and laboratory variables are included (green circle). Unlike the CHRS model, the MN-predict model provides risks for specific categories of myeloid malignancy. For my patient, the model indicated a high-risk of developing MDS or CMML, with lower risks of developing MPN or AML. Abbreviations: variant allele fraction (VAF); mean corpuscular volume (MCV); red cell distribution width (RDW); hemoglobin; PLT, platelet count (HGB); absolute neutrophil count (ANC); reticulocytes (Retic); mean platelet volume (MPV); platelet distribution width (PDW); body mass index (BMI).
The CHRS model uses input variables (blue circle) to calculate the risk of developing any myeloid malignancy and predict overall survival. Inputting the variables from my patient indicated a high-risk assignment. The MN-predict model uses similar input variables as the CHRS model, although several additional clinical and laboratory variables are included (green circle). Unlike the CHRS model, the MN-predict model provides risks for specific categories of myeloid malignancy. For my patient, the model indicated a high-risk of developing MDS or CMML, with lower risks of developing MPN or AML. Abbreviations: variant allele fraction (VAF); mean corpuscular volume (MCV); red cell distribution width (RDW); hemoglobin; PLT, platelet count (HGB); absolute neutrophil count (ANC); reticulocytes (Retic); mean platelet volume (MPV); platelet distribution width (PDW); body mass index (BMI).
Competing Interests
Dr. Hasserjian indicated no relevant conflicts of interest.