The bone marrow is a rough-and-tumble place. As we age, our hematopoietic stem cells (HSC) progressively accumulate mutations, some of which confer a survival advantage that allows HSC clones to expand and outcompete nonmutated HSCs within the bone marrow.1 When genetic studies such as next-generation sequencing (NGS) detect these telltale clonal mutations in blood or bone marrow in patients without cytopenias or hematologic malignancy, this process is termed clonal hematopoiesis of indeterminate potential (CHIP).
In CHIP, a growth-advantaged HSC clone contributes to the production of more blood cells than its non-mutated neighbors. CHIP is identified by the detection of a mutation in a gene associated with myeloid neoplasia, at a variant allele frequency (VAF) of at least 2% in blood or bone marrow.2 Genes most commonly mutated in CHIP are those that regulate DNA methylation and RNA splicing, which contribute to the affected HSC’s enhanced fitness in the aging marrow microenvironment. Individuals with CHIP have an increased risk of developing hematologic neoplasms, as well as increased mortality due to vascular disease.3 Since the rate of mutation acquisition in HSCs appears to be constant,1 it is of great interest to understand what drives the expansion of CHIP clones in individual patients — a key factor in the development of hematologic malignancy and mortality.4 One well-known factor is exposure to cytotoxic therapies, which select for and promote the expansion of preexisting CHIP clones that bear specific mutations in DNA damage response genes (such as TP53 and PPM1D).5 However, these environmental factors likely do not fully explain the highly variable destiny of CHIP clones in individuals.
Dr. Weinstock and colleagues peered into the genetic signatures of HSC clones using a technique called PACER (passenger-approximated clonal expansion rate). The PACER method is based on the fact that the “driver” mutation that confers the growth advantage to the HSC clone is accompanied by “passenger” mutations that the HSC acquired prior to acquiring the driver mutation. A higher number of passenger mutations indicates that the clone arose later in life, and thus expanded to the detected size more quickly and would be expected to continue to grow quickly in the future. The yearly growth rate of HSC CHIP clones (compared with that of non-mutated HSCs) reflects the clones’ fitness in each individual. Clone growth in turn influences the risk of adverse consequences such as development of a myeloid malignancy. The authors validated PACER’s ability to predict future clone growth by sequencing a cohort of 55 CHIP carriers who had two separate blood samples taken over time and found that PACER accurately estimated the clone’s expansion over time in each individual.
The authors applied PACER to estimate the growth rate of clones in 4,536 individuals who carried CHIP clones from a TOPMed population study program. They then performed a genome-wide association study to identify germline genetic variants that correlated with the CHIP clone growth rate. The authors identified a variant in the TCL1A gene that was significantly associated with altered growth of CHIP clones. Specifically, the rs2887399 variant (representing a T to G substitution in the core promoter of TCL1A) was associated with decreased CHIP clone growth; this variant was present in 26% of individuals in the TOPMed cohort. For example, TET2 and ASXL1 clones expanded by a predicted rate of 8.3% per year in individuals with the typical T/T rs2887399 genotype, compared with a rate of only 0.5% per year in individuals with the homozygous G/G rs2887399 variant genotype. The rs2887399 variant was also associated with decreased incidence of CHIP mutations in general, likely due to their slowed expansion up to the 2% VAF level that defines CHIP.
In Brief
The identification of a germline variant in TCL1A affecting expansion of an HSC clone is surprising, since TCL1A is not a known myeloid oncogene and has not been previously implicated in myeloid malignancy. The TCL1A protein encoded by this gene is expressed in B cells and plasmacytoid dendritic cells, but not in other hematopoietic cell types. By examining single-cell RNA sequencing databases, the authors found that, indeed, TCL1A was not expressed significantly in HSCs in normal marrow; however, it was expressed more frequently in HSCs from patients with myeloid malignancies bearing TET2 or ASXL1 mutations. The work of Dr. Weinstock and colleagues suggests a model where the TCL1A gene is normally repressed in HSCs but is aberrantly expressed in upon acquisition of CHIP mutations such as TET2. In turn, this aberrant expression drives clonal expansion of the affected HSCs. The rs2887399 variant in the TCL1A promoter down-regulates TCL1A protein expression, thus slowing expansion of the clone relative to individuals lacking this variant (Figure). The authors explored this model in murine and in vitro models of hematopoiesis and showed that forced TCL1A expression in HSCs increased their proliferative cycling, promoted their expansion, and fostered their selective engraftment in murine bone marrow — truly enhancing the fitness of the clone relative to that of TCL1A non-expressing HSCs.
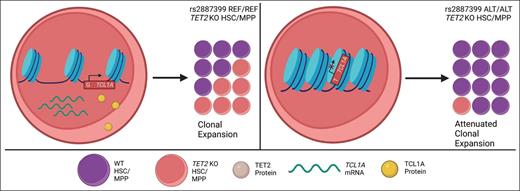
Proposed model of the interaction of TCL1A expression with TET2 CHIP clonal expansion. In TET2-mutated HSC clones, TCL1A protein expression is enhanced due to increased TCL1A promoter accessibility, promoting expansion of the clone (left panel). The germline rs2887399 TCL1A variant limits promoter accessibility, abrogating increased TCL1A protein expression and reducing expansion of the TET2 clone (right panel). Reprinted with permission from Springer Nature. Weinstock JS et al. Nature. 2023;616(7958):755-763.
Proposed model of the interaction of TCL1A expression with TET2 CHIP clonal expansion. In TET2-mutated HSC clones, TCL1A protein expression is enhanced due to increased TCL1A promoter accessibility, promoting expansion of the clone (left panel). The germline rs2887399 TCL1A variant limits promoter accessibility, abrogating increased TCL1A protein expression and reducing expansion of the TET2 clone (right panel). Reprinted with permission from Springer Nature. Weinstock JS et al. Nature. 2023;616(7958):755-763.
The work of Dr. Weinstock and coauthors has added a germline predisposition twist to the implications of acquired CHIP mutations. An inherited germline variant in the promoter of the TCL1A gene — present in about one-quarter of the population — appears to slow the expansion of mutated HSC clones, thereby both suppressing the development of CHIP and slowing the increase in the size of CHIP clones. This model indicates that genetic variation within individuals cooperates with environmental factors (such as exposure to cytotoxic therapy) in the marrow response to aging, often fostering the emergence of maladaptive mutated HSC clones that contribute to risk of hematologic malignancy and cardiovascular disease. The authors speculate that pharmacologic manipulation of TCL1A expression could be harnessed to treat (or prevent the development of) hematologic malignancies related to CHIP gene mutations. However, TCL1A expression may also play a role in the adaptation of stem cells to the aging marrow microenvironment and marrow stress response. Further study is needed to determine the downstream effects of its suppression.
Competing Interests
Dr. Hasserjian reports receiving consulting income from AstraZeneca, Bluebird Bio, Daiichi Sankyo, and Jazz Pharmaceuticals.