The current World Health Organization (WHO) classification of myeloid neoplasms defines disease groups based on the percentage of blood and marrow blasts, with cases presenting with at least 20 percent blasts classified as acute myeloid leukemia (AML), and additionally subclassified according to specific cytogenetic abnormalities, mutations (e.g., NPM1, RUNX1, or bi-allelic CEBPA), and disease ontogeny (e.g., therapy-relatedness). The TP53 gene is a critical component of the DNA damage response, and mutation in this gene confers an aggressive phenotype to myelodysplastic syndromes (MDS), AML, and numerous other hematopoietic and non-hematopoietic neoplasms.1,2 TP53 mutation is associated with genetic instability and a complex karyotype. While the latter finding defines AML with myelodysplasia-related changes (AML-MRC) in the current WHO classification, neither TP53 mutation nor complex karyotype influence the classification of MDS. Recent studies have suggested that from a molecular standpoint, MDS and AML may be artificially segregated from one another based on the historic 20 percent blast cutoff.3 With respect to TP53 mutation, it bears exploring whether TP53 mutation, rather than a specific blast percentage or traditional disease-defining cytogenetic features, should define a unified disease group.
Dr. Tim Grob and colleagues performed a comprehensive analysis on 2,200 patients with AML and MDS with excess blasts (MDS-EB) who received standard induction chemotherapy and consolidation according to Haemato-Oncology Foundation for Adults in the Netherlands and Swiss Group for Clinical Cancer Research (HOVON-SAKK) protocols, focusing on patients who had TP53 mutations detected by next-generation sequencing (NGS). They found TP53 mutations in 230 patients (10.5% of the entire study group, including 186 AML and 44 MDS-EB patients). The TP53 mutations included missense (72.7%), nonsense (5.6%), insertion/deletion (13.4%), and splice-site (8.1%) mutations. All missense mutations occurred in the TP53 domain responsible for binding to DNA, leading to a loss of function and preventing transcriptional activation of gene targets. Overall, 75.7 percent of the TP53 mutations were multi-hit, reflecting an abnormality of the second TP53 allele by mutation, deletion, or loss of heterozygosity. Concurrent co-mutations were detected in 49 percent of the mutant TP53 cases, with the most frequently associated mutations being DNMT3A, TET2, ASXL1, and SRSF2.
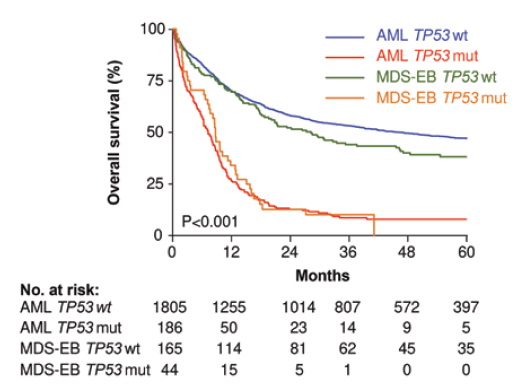
Consistent with prior studies, Dr. Grob and colleagues found that mutant TP53 was associated with very poor survival (2-year overall survival [OS], 12.8%), which was significantly worse compared to other patients with European LeukemiaNet (ELN) adverse-risk TP53–wild-type AML in their cohort (2-year OS, 42.5%; p<0.001). Interestingly, the authors did not find any significant difference in clinical outcome based on the TP53 mutant allelic status (multi-hit vs. a single TP53 mutation with an intact TP53 wild-type allele), TP53 clone size, presence of specific co-mutations, and WHO Classification as AML versus MDS-EB (Figure). Complex karyotype was seen in 84 percent of the TP53-mutated cases in their series and was even further enriched in the TP53 multi-hit cases. Of note, the small number of TP53-mutated patients with non-complex karyotype did have superior survival to those with complex karyotype (2-year OS, 34% vs. 9%, respectively, p=0.002). However, importantly, among complex karyotype patients, TP53 conferred a significantly worse outcome.
Overall survival of myelodysplastic syndrome with excess blasts (MDS-EB) and acute myeloid leukemia (AML) at diagnosis subclassified by wild-type TP53 and mutated TP53 cases. Patient survival in this cohort is stratified by the presence or absence of TP53 mutation, not classification as MDS-EB versus AML (modified from Figure 2B, Grob et al. Blood. 2022).
Overall survival of myelodysplastic syndrome with excess blasts (MDS-EB) and acute myeloid leukemia (AML) at diagnosis subclassified by wild-type TP53 and mutated TP53 cases. Patient survival in this cohort is stratified by the presence or absence of TP53 mutation, not classification as MDS-EB versus AML (modified from Figure 2B, Grob et al. Blood. 2022).
Finally, the authors performed deep targeted sequencing analysis on bone marrow samples of 62 patients with TP53-mutated AML in complete remission to assess for molecular minimal residual disease (MRD), which has been shown to correlate with outcome in some AML subtypes.4 They found detectable TP53 mutations indicating MRD in 45 of 62 (72.5%) of the remission marrow samples. There was no difference in OS between patients with and without detectable TP53 mutation in remission (p=0.653).
The study by Dr. Grob and colleagues shows that TP53 mutation is a highly adverse feature that penetrates all conventional boundaries used to classify and risk-stratify MDS and AML. TP53 mutations are currently placed in the adverse risk category of the AML ELN 2017 scheme.5 However, the data presented by Dr. Grob and colleagues indicate that TP53-mutated AML has particularly poor outcomes that set it apart from other adverse-risk AML, including those with complex karyotype and wild-type TP53. The study also did not show a significant difference in outcome (or in co-mutation profiles) between MDS-EB and AML with mutant TP53 when treated similarly, suggesting that these diseases may be inappropriately segregated from one another based on the 20 percent blast cutoff. The overall similarity of AML/MDS-EB with mono-allelic versus multi-hit TP53 mutation contrasts with the differences of single versus multi-hit TP53 mutation in MDS reported by Dr. Elsa Bernard and colleagues.6 One explanation for this discrepancy could be the exclusion of non-excess blast MDS cases from the study by Dr. Grob and colleagues; it is possible that the outcome of lower-risk MDS cases is more affected by the TP53 allelic status than those with higher blast counts.7
In Brief
The work by Dr. Grob and colleagues has demonstrated that in a large cohort of intensively treated patients, TP53-mutated MDS-EB and AML share biologic features and clinical behavior and collectively represent a highly aggressive disease group. Unlike some other AML subtypes, MRD assessment in remission does not appear to predict outcome in TP53-mutated AML. The dismal outcome of this “ultra-high–risk” tranche of MDS and AML warrants novel treatment approaches that focus on the TP53-mutated state rather than allocation to traditional MDS or AML therapies based on which side of 20 percent the blast count falls.
Competing Interests
Dr. Ta and Dr. Hasserjian indicated no relevant conflicts of interest.