Genetic analysis is an integral part of the diagnosis and subclassification of acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS), and powerfully impacts clinical risk-stratification of these diseases. The 2017 European Leukemia Network (ELN) and 2012 Revised International Prognostic Scoring System (IPSS-R) incorporate several cytogenetic aberrations and, in the case of the ELN, mutations in certain genes to stratify patients. Timely and accurate assignment of newly diagnosed patients to the appropriate risk group is critical in directing patient management. Moreover, several newly approved AML therapies are indicated in the context of certain cytogenetic findings such as the use of CPX-351 (daunorubicin-cytarabine) in AML with myelodysplasia-related changes based on complex cytogenetic aberrations, or gemtuzumab ozogamicin in core-binding factor AML with inv(16)/t(16;16) or t(8;21).1 The current “gold standard” method for detecting cytogenetic aberrations is to grow leukemic cells in culture and examine the metaphases of dividing cells (conventional karyotyping). While conventional karyotyping comprehensively analyzes gross changes in the genome (chromosomal rearrangements, gains, and losses), it can be limited by failure of cells to grow in culture, as well as so-called “cryptic” abnormalities in which key translocations are not visualized on metaphase preparations.2 Moreover, the turnaround time for conventional karyotyping of bone marrow can be up to 10 days and is longer in some labs, potentially introducing a significant delay in up-front chemotherapy. Fluorescence in situ hybridization (FISH) abrogates some of these limitations, but this requires the application of specific probes and also may be subject to false negative or false positive results.3
Whole-genome sequencing (WGS) comprehensively examines the entire genome and can detect a wide spectrum of genetic alterations, ranging from gross genetic changes visible in conventional karyotyping to changes in single nucleotides. Unlike conventional karyotyping, WGS can be performed on nonviable cells, including fixed and paraffin-embedded material. Dr. Eric Duncavage and colleagues recently applied WGS to 263 newly diagnosed patients with AML and MDS. The WGS was designed to detect single gene mutations, genome-wide copy-number alterations, and structural chromosomal variants found in MDS and AML. The WGS results were used to risk stratify patients with AML and MDS according to ELN and IPSS-R schemes, respectively, and were compared to the risk assignments based on standard methods of conventional karyotyping combined with next-generation sequencing (NGS). In a head-to-head comparison, the sensitivity of WGS to detect recurrent cytogenetic translocations and copy-number alterations was 100 percent; additionally, WGS detected 13 cytogenetically cryptic structural variants and 21 copy-number alterations that were missed on karyotype. The sensitivities of WGS to detect point mutations and insertion-deletion mutations were 84.6 percent and 91.5 percent, respectively. This lower sensitivity to detect mutations compared to standard NGS panels likely reflects lower depth of coverage of WGS (mean, 50×) compared to targeted NGS panels (>500×), causing some subclonal variants present at low variant allele fraction (VAF) to be missed. Overall, compared to conventional genetic testing, WGS provided new genetic information in 25 percent of patients. Moreover, 15 percent of the patients with non-acute promyelocytic leukemia AML and 21 percent of the patients with MDS were assigned to different risk categories based on the WGS results.
The results of the study by Dr. Duncavage and colleagues are tremendously exciting for those who diagnose and treat patients with MDS and AML for several reasons:
WGS improved the overall accuracy of genetic testing by providing results in cases in which conventional karyotype failed, were ambiguous, or missed significant findings. WGS-based testing provided more significant ELN risk stratification (p=0.01) compared to the conventional genetic technologies (p=0.09) in 71 patients with AML with outcome information, and successfully risk-stratified 27 patients with AML in which cytogenetics failed or yielded inconclusive results. The abnormalities detected by WGS included findings such as core-binding factor translocations or complex karyotypes that may have changed the therapeutic approach.
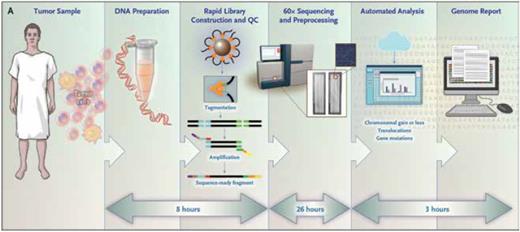
WGS represents a “one-stop-shop” test that gives all the genetic information (except for FLT3-ITD assessment, which was run as a separate polymerase chain reaction [PCR] –based test) needed for diagnosis and risk-stratification (Figure). This would simplify the current workflow that requires submission of separate aliquots of bone marrow for conventional karyotyping and NGS, and in some cases, FISH analyses, RNA-based fusion assays, and reverse transcription-PCR–based translocation detection. It also obviates the need to provide viable leukemic cells for culture and can be performed on smaller samples than are typically required for conventional karyotyping.
The average turnaround time for WGS (from sample submission to results) was 5.1 days, which represents a significant improvement over typical turnaround times for conventional karyotype and clinical NGS panels. This would allow more rapid final diagnosis and institution of appropriate therapy that is informed by the genetic risk.
Workflow and approximate processing time for each step of the rapid whole-genome sequencing method used for samples obtained from the study patients. QC, quality control. From N Engl J Med, Duncavage EJ, Schroeder M, O'Laughlin M, et al, Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers, vol. 384, pp 924-935, Copyright © 2021 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.
Workflow and approximate processing time for each step of the rapid whole-genome sequencing method used for samples obtained from the study patients. QC, quality control. From N Engl J Med, Duncavage EJ, Schroeder M, O'Laughlin M, et al, Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers, vol. 384, pp 924-935, Copyright © 2021 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.
In Brief
One limitation is the lower sensitivity of WGS to detect subclonal mutations compared to standard NGS panels, which have higher depth of coverage. Despite missing 12.5 percent of mutations that impact ELN classification, the positive predictive value of the WGS assay to detect variants at a VAF of at least 5 percent was more than 99 percent; further study is needed to determine the contribution of low-level subclonal mutations to patient outcome in MDS and AML. Additionally, the cost of WGS has been generally considered to be prohibitive for use in routine clinical testing. However, due to the continual decrease in the cost of sequencing through advancing technologies, the authors estimate that the cost of applying WGS in a high-volume laboratory could be as low as $1,300, which is within the range of the cost of current genetic testing applied to AML and MDS diagnosis.4 This novel approach to comprehensive genomic analysis of myeloid neoplasms is thus already visible on the horizon and could help streamline the currently complex and variable genetic testing algorithms.
Competing Interests
Dr. Hasserjian indicated no relevant conflicts of interest.