2019 marked a great leap forward in our molecular understanding of chronic myeloid neoplasms (CMNs), with results that may challenge long-held views on how to classify and diagnose them. CMNs comprise a diverse group of malignancies characterized by an abnormal proliferation of maturing hematopoietic cells, morphologic dysplasia, or a combination of both. Traditional classification of these disorders based solely upon clinicomorphologic observations has resulted in heterogeneity within diagnostic groups and poor patient outcomes.1,2 However, as advances in genomics expand our awareness of their biology, genetic lesions are increasingly required as primary diagnostic criteria within the World Health Organization (WHO) classification system (particularly in myeloproliferative neoplasms [MPNs]) and often provide targets for therapeutic intervention in CMNs.3 A growing body of literature also demonstrates that the clonal architecture of additional mutations acquired by many CMNs can provide actionable prognostic information.2,4 Nevertheless, progress has not been made evenly across all chronic myeloid neoplasms. With the exception of chronic myeloid leukemia, myeloid neoplasms presenting predominantly with neutrophilia represent a relatively underexplored area of genomic investigation.
Myeloid neoplasms presenting predominantly with neutrophilia (henceforth, neutrophilic myeloid neoplasms [NMNs]) include chronic neutrophilic leukemia (CNL), atypical chronic myeloid leukemia (aCML), myelodysplastic syndromes (MDSs)/MPNs, and unclassifiable MDSs/MPNs (MDS/MPN-U). Several challenges persist in characterizing these neoplasms effectively. First, as a group, the disorders are rare, inhibiting the large-scale genomic analyses or clinical trials afforded to other CMNs. Second, there is no gold standard for diagnosis or discrimination of these entities. Patients may present with clinical and morphologic features insufficient for diagnosis or may fulfill criteria for multiple diagnoses simultaneously. The presence of distinguishing features such as morphologic dysplasia, left-shifted granulocytic maturation, or monocytosis may be confounded by other clinical factors or be subject to inter-pathologist variation. Initial genomic investigations into these disorders, often using smaller targeted panels, yielded recurrent mutations across all these entities in genes such as ASXL1, TET2, SRSF2, CBL, CSF3R, SETBP1, NRAS, U2AF1, and EZH2, among others, in common with many other myeloid neoplasms.5 The question remained whether molecular distinctions between these entities might be more apparent with a more genome-wide investigation.
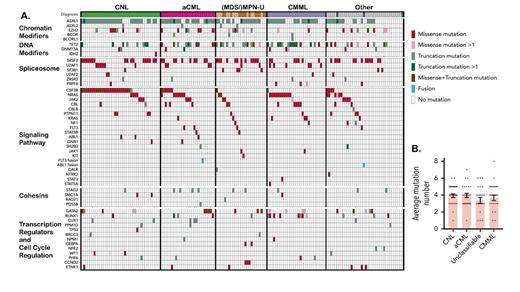
(A) Mosaic plot exhibiting recurrent mutations categorized by clinicopathologic diagnosis and pathway (adapted with permission from Zhang H et al. Blood. 2019;134:867-879). (B) Average number of mutations per patient (reproduced with permission from Zhang H et al).
(A) Mosaic plot exhibiting recurrent mutations categorized by clinicopathologic diagnosis and pathway (adapted with permission from Zhang H et al. Blood. 2019;134:867-879). (B) Average number of mutations per patient (reproduced with permission from Zhang H et al).
To address this question, Dr. Haijiao Zhang and colleagues performed whole-exome sequencing of bone marrow and/or peripheral blood samples from 158 patients with NMN diagnoses (Figure 1).6 Specimens included 39 cases of CNL, 27 cases of aCML, 13 cases of MPN-U, 12 cases of MDS/MPN-U, 29 cases of chronic myelomonocytic leukemia (CMML), and 38 cases of uncertain/unavailable diagnosis. It should be noted that these samples originated from a large number of institutions without central pathology review for these challenging diagnoses. In keeping with previous studies on all CMNs,5 the researchers detected high rates of mutations in genes that fall within only a few key pathways: chromatin modification (ASXL1, EZH2, ASXL2), signaling pathway driver mutations (CSF3R, NRAS, JAK2), DNA modifier mutations (TET2, DNMT3A), spliceosome component mutations (SRSF2, U2AF1, SF3B1, ZRSR2, RPRF8), and transcription factor mutations (GATA2, RUNX1; Figure 1). Additionally, there was a high frequency of SETBP1 mutations, though mutations in this gene may be seen, especially as progression events, in other CMNs as well. Three or more of these major pathways were represented in most cases (50% containing 3 or 4 mutations; range, 0-8 mutations) with mutation profiles most frequently including ASXL1/2, TET2/GATA2 (mutually exclusive), a signaling mediator, and/or a splicing factor. Analysis of variant allele fractions allowed the authors to infer that variants in EZH2, SETPB1, TET2, U2AF1, and SF3B1 (chromatin modifier and spliceosome pathways) represented the most frequent founder clones, whereas other mutations occurred at more variable frequencies, with evidence of linear acquisition of mutations. Loss of heterozygosity was observed frequently with mutations in EZH2, JAK2, GATA2, and CSF3R.
Importantly, the authors detected no definitive segregation of mutational profiles between CNL, aCML, and other NMN diagnoses as defined by WHO criteria applied at the individual institutions. RNA-seq analyses performed on 76 patient samples revealed three main transcriptomic clusters that enriched for, but also did not absolutely discriminate, clinical or genomic disease features. This indicates that NMN diagnostic entities do not exhibit distinct molecular profiles from each other; however, the collection may comprise a larger entity from which a spectrum of clinicomorphologic manifestations can arise. Of note, a subset of mutations including NRAS, ASXL1, GATA2, and DNMT3A were significantly associated with poorer patient survival irrespective of clinicomorphologic diagnosis within the NMN group while mutations in CBL showed improved outcomes in univariate analysis, suggesting that molecular findings may represent more clinically actionable information than the specific diagnostic classifications within the NMNs.
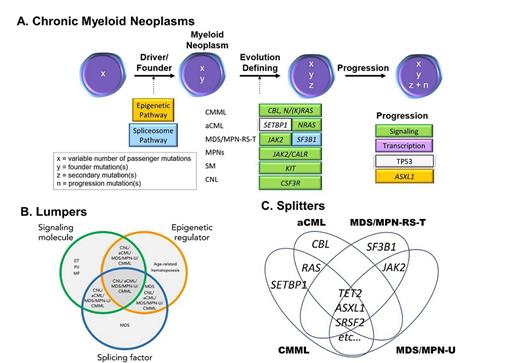
(A) Schematic of common changes acquired in the differentiation and progression of chronic myeloid neoplasms (adapted from McClure RF et al. J Mol Diagn. 2018;20:717-737). (B) Lumpers: comparison of the NMNs with myeloproliferative neoplasms (MPNs), myelodysplastic syndromes (MDS), and age-related clonal hematopoiesis (adapted from Zhang H et al. Blood. 2019;134:867-879). (C) Splitters: comparison between the NMNs (diagram derived from Meggendorfer M et al. Haematologica. 2018 May;103:e192-e195). CNL, chronic myeloid neoplasms; aCML, atypical chronic myeloid leukemia; CMML, chronic myelomonocytic leukemia; CNL, chronic neutrophilic leukemia; ET, essential thrombocythemia; MF, myelofibrosis; MPN-RS-T, MPN with ring sideroblasts and thrombocytosis; MPN-U, unclassifiable MPN; PV, polycythemia vera; SM, systemic mastocytosis.
(A) Schematic of common changes acquired in the differentiation and progression of chronic myeloid neoplasms (adapted from McClure RF et al. J Mol Diagn. 2018;20:717-737). (B) Lumpers: comparison of the NMNs with myeloproliferative neoplasms (MPNs), myelodysplastic syndromes (MDS), and age-related clonal hematopoiesis (adapted from Zhang H et al. Blood. 2019;134:867-879). (C) Splitters: comparison between the NMNs (diagram derived from Meggendorfer M et al. Haematologica. 2018 May;103:e192-e195). CNL, chronic myeloid neoplasms; aCML, atypical chronic myeloid leukemia; CMML, chronic myelomonocytic leukemia; CNL, chronic neutrophilic leukemia; ET, essential thrombocythemia; MF, myelofibrosis; MPN-RS-T, MPN with ring sideroblasts and thrombocytosis; MPN-U, unclassifiable MPN; PV, polycythemia vera; SM, systemic mastocytosis.
In addition to challenging the clinicomorphologic boundaries currently held by WHO criteria, the mutational profiles detected within NMNs conform to the larger schema of pathogenic mutation acquisition established for CMNs writ large (Figure 2A).5 This schema holds that initiating/founder events in CMNs are predominated by mutations involving epigenetic regulators and/or RNA splicing machinery. In CMNs other than MDS, a secondary mutational event(s) defines the differentiation and/or manifestation of the CMN, including JAK2, CALR, or MPL for “classic” MPNs. The recent work by Dr. Haijiao Zhang and colleagues suggests NMNs are subject to this second “hit” through a variety of mutations, often signaling pathway genes (frequently co-occurring) that “may contribute to the unique phenotype” of these NMNs, but not in a manner that reliably distinguishes them from each other. Lastly, increased numbers of mutations are associated with worse clinical features progression. Thus, it seems that neutrophilic neoplasms obey a similar general strategy of pathogenic progression, while sharing skewed representation of individual genes within these categories.
Notably, these results stand in contrast to two publications by Dr. Manja Meggendorfer and colleagues, performed with comparable sample sizes to those from Dr. Haijiao Zhang and colleagues.7,8 The studies by Dr. Meggendorfer and collaborators hold that the frequency of specific gene mutations may help discriminate amongst the NMNs (Figure 2B). Their data suggest that mutations in SETBP1 are significantly enriched in aCML, whereas mutations in CBL in the context of monocytosis suggest a diagnosis of CMML. Both CMML and aCML share an enrichment for RAS pathway mutations while MDS/MPN-U and MDS/MPN with ring sideroblasts and thrombocytosis share increased frequency of JAK2 mutations. While enrichments for particular mutations may be seen in retrospective analyses, the danger of this approach from a diagnostic perspective is that all of the genes may be mutated in other entities as well.
Collectively, the findings from Dr. Zhang and colleagues challenge us to reconsider CMNs presenting predominantly with neutrophilia, including CNL, aCML, and MDS/MPN-U, as a spectrum of manifestations arising from a common set of genetic mutations (Figure 2C). This prompts two important questions: Are molecular constellations more prognostically useful than current clinicomorphologic diagnostic categories? Even more radically, do we need morphologic distinctions between the NMNs, especially ones that are poorly reproducible and challenging, in this new genomic era? While hematologic parameters and morphologic evaluation still comprise essential and predictive components of the diagnosis of neutrophilic neoplasms at present, it is difficult to envision a future in which genomics and other molecular analyses do not play an increasingly predominant role in governing the classification and management of these disorders. In one such example, Dr. Zhang and coauthors, and Dr. Robert Hasserjian in his associated commentary,9 have questioned whether CNL, technically categorized as an MPN, is not more molecularly similar to aCML and MDS/MPN-U and therefore more appropriately grouped with them. Moving forward, as in the case of the MPNs,4 we may discover new subcategories for NMNs defined by mutational architecture and orthogonal to traditional morphologic categories, or perhaps a more inclusive understanding of epigenomic, proteomic, and other changes will illuminate individualized presentations, clinical courses, and appropriate therapies in the future.
References
Competing Interests
Dr. Hergott and Dr. Kim indicated no relevant conflicts of interest.