Myeloproliferative neoplasms (MPNs) arise through clonal expansion of hematopoietic stem cells (HSCs) with oncogenic mutations in JAK2 , MPL , or CALR , leading to constitutive activation of the JAK-STAT pathway. This apparently simple genetic driver landscape of MPNs belies the diverse clinical presentations, which vary in the expanded differentiated myeloid population, leading to essential thrombocythemia (ET), polycythemia vera (PV), or primary myelofibrosis (PMF). Cytotoxic treatment with hydroxyurea or targeted inhibition of JAK2 controls disease symptoms; however, these diseases have substantial implication on quality of life and increased risk for thrombosis, progression to myelofibrosis, or acute myeloid leukemia.
The clinical discordance between common driver mutations and MPN phenotype has been attributed to host factors, regulation of stem cells within the microenvironment (e.g., aberrant cytokine signaling), or the acquisition of secondary mutations (e.g., in epigenetic regulators or TP53). Even within individual patients, there may be multiple MPN clones coexisting, suggesting a stepwise evolution of MPN-pathogenesis. Emerging sequencing technologies enable characterization of single cells of heterogeneous cell populations through their transcriptome, but technical limitations have hampered the simultaneous detection of somatic mutations. A major challenge within the field has been to link the genotype of a single cell to its transcriptional phenotype and therefore to understand the factors that regulate clinical heterogeneity and disease progression.
To overcome this barrier, Dr. Alba Rodriguez-Meira and colleagues developed TARGET-seq. Their innovative method increases single-cell RNA sequencing library complexity, and an optimised lysis allows for the targeted amplification of multiple loci in both genomic DNA (gDNA) and complementary DNA (cDNA). Genotyping on both gDNA and cDNA increased the sensitivity to detect somatic mutations in single cells to more than 98 percent, also overcoming allelic dropouts (ADO) of low or not-expressed transcripts.
They performed this high-resolution parallel single-cell genotyping and transcriptomics on samples from patients with MPN in whom there were different combinations of frequently occurring somatic mutations in epigenetic modifiers ( EZH2 , TET2 , and ASXL1 ). TARGET-seq on CD34 + HSCs allowed inference of the order of mutation acquisition to reveal cancer evolution. They found that TET2 mutation was an early event and was present prior to acquisition of disease-driving mutation JAK2V617F . This is in line with findings that mutations in epigenetic modifiers lead to clonal haematopoiesis (CH) and increases the risk of developing blood malignancies. Contrarily, CH-associated mutations in ASXL1 were acquired late. Notably, TARGET-seq confidently discriminated between single cells with heterozygous or homozygous JAK2V617F mutations. The ability to recover mutant-gene dosage is of considerable importance as it informs prognosis in MPNs and many other cancers.
Little is known about how the combination of mutations can alter a cell’s transcriptional program and the microenvironment. Solely looking at the transcriptomic cellular identity using T-distributed Stochastic Neighbor Embedding (t-SNE), Dr. Rodrigues-Meira and colleagues identified that the MPN and wild-type HSCs shared many transcriptional features characterized by increased inflammatory response and activated MYC targets compared with healthy donor HSCs. This is the first time that alterations in normal HSC function have been observed in patients with MPN and suggests that additional cell extrinsic factors, such as the microenvironment, are at play and drive a pathological transcriptional phenotype in cells even when they lack MPN mutations.
Globally, wild-type and mutant MPN HSC could not be separated; however, TARGET-seq’s high-resolution genotyping enabled comparison between wildtype and mutant cells in the same patient, allowing an unbiased assessment of a mutation’s cell intrinsic effects. They found dysregulation of STAT5A targets, activation of oncogene MYCN , and tumor suppressor TP53 , consistent with described effects linked to JAK2V617F biology. Cells with lesions in epigenetic modifiers EZH2 or TET2 showed distinct, mutation-specific transcriptional changes. By comparing mutant with wild-type HSCs, it was possible to identify potential biomarkers or therapeutic targets shared for PV and PMF.
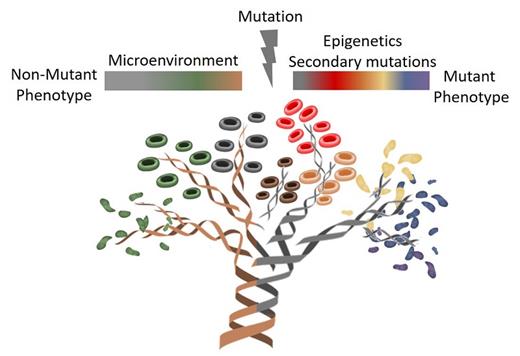
Combined single-cell genotyping and transcriptional analysis using TARGET-seq enables researchers to discriminate wild-type and heterogeneous mutated cells within individual patients with MPN. This allowed characterization of mutation-specific and nonmutant transcriptional phenotypes, and points toward mediators of clinical manifestations and progression. Illustration by Jasmin Straube, PhD, QIMR Berghofer, Herston, Australia.
Combined single-cell genotyping and transcriptional analysis using TARGET-seq enables researchers to discriminate wild-type and heterogeneous mutated cells within individual patients with MPN. This allowed characterization of mutation-specific and nonmutant transcriptional phenotypes, and points toward mediators of clinical manifestations and progression. Illustration by Jasmin Straube, PhD, QIMR Berghofer, Herston, Australia.
In this study, the Mead lab has developed a novel technology to simultaneously characterize the genotype and phenotype of a heterogeneous population of cancer cells, providing a powerful tool to understand the evolution of disease progression (Figure). The broad applicability and importance of this approach is further evidenced by recent publications from Dr. Anna S. Nam and colleagues 1 and Dr. Peter van Galen and colleagues, 2 who have used different approaches that rely on genotyping cDNA. Using dual gDNA and cDNA genotyping, the approach of Dr. Rodriguez-Meiras’ team robustly discriminates heterozygous from homozygous mutations, detects loss of heterozygosity, and reduces ADO without losing transcriptomic information. A limitation of these techniques is that genotyping done through targeted amplification is not suitable for mutation discovery. Furthermore, these techniques are labor and resource sensitive and are therefore currently not suitable for routine clinical use. A further challenge is dissecting out the clear transcriptional effects of treatments such as interferon-α or JAK-inhibitors.
In Brief
Overall, this work highlights the complexity of MPNs and stresses the importance of the integration of multi-omics approaches to understand disease biology and to identify effective therapeutic targets. This article has generated a wealth of data adding to the growing number of resources readily available for hypothesis generation and testing, as well as validation of murine or in vitro models of disease, and will enhance the capability of the research community to target MPNs.
References
Competing Interests
Dr. Straube and Dr. Lane indicated no relevant conflicts of interest.