The phenomenal success achieved in the treatment of pediatric acute lymphoblastic leukemia (ALL) stands out as one of the great achievements in cancer medicine.1 Unfortunately, a small minority of patients will still have disease that is refractory to initial chemotherapy, or disease that relapses after chemotherapy, and these patients continue to have poor long-term outcomes. Glucocorticoids are an essential component of chemotherapy in lymphoid malignancies (ALL, lymphoma, and myeloma) but have limited efficacy in myeloid, or indeed in many other cancers. This discrepancy suggests a conserved mechanism of tissue-specific response to glucocorticoids. It follows therefore that if such a mechanism exists, and this is independent of the genomic factors giving rise to leukemia, there may also be a common, nongenomic process that regulates acquired glucocorticoid resistance in ALL. These intriguing observations linking tissue specificity of glucocorticoid response and acquired glucocorticoid resistance were examined in a recent article by Dr. Duohui Jing and colleagues.
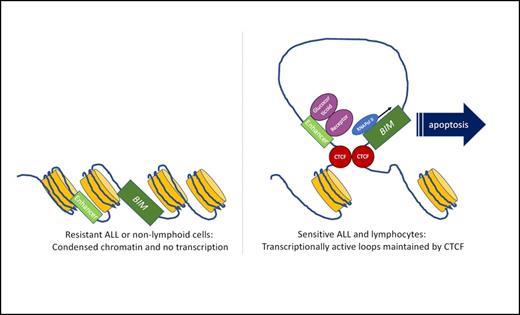
Proposed model of glucocorticoid sensitivity. Open chromatin loops facilitate interactions between enhancer regions and effector molecules such as the proapoptotic protein, BIM. In nonlymphoid cells, or resistant ALL, this chromatin is inaccessible, which prevents downstream transcriptional activation of glucocorticoid targets.
Proposed model of glucocorticoid sensitivity. Open chromatin loops facilitate interactions between enhancer regions and effector molecules such as the proapoptotic protein, BIM. In nonlymphoid cells, or resistant ALL, this chromatin is inaccessible, which prevents downstream transcriptional activation of glucocorticoid targets.
Chromatin conformation mediates many biologic functions, including cellular differentiation, by facilitating DNA accessibility of key lineage-specific transcription factors and regulators. Each cell type has 70 to 100,000 accessible chromatin domains that interact with transcriptional regulators to generate a complex interactive network that is highly correlated with cellular identity, a kind of three-dimensional cellular fingerprint. Using a bioinformatics approach and in silico data, the authors identified a specific chromatin conformation pattern found in lymphocytes. Interestingly, although some of the open, transcriptionally active sites correlated with lymphoid transcription factors, many were related to other cellular processes including hematopoietic development and the induction of apoptosis. When the authors examined glucocorticoid receptor binding sites specifically, they observed that these were highly correlated with open chromatin in lymphocytes, and closed chromatin in myeloid cells. Additionally, CTCF, a chromatin architectural protein that maintains DNA loops, was specifically enriched at the areas of open chromatin that also had glucocorticoid receptor binding, suggesting that this genomic architecture was a prerequisite for glucocorticoid receptor–mediated transcriptional changes and consequent downstream signalling. In particular, they identified chromatin accessibility at regulatory regions associated with the proapoptotic factor, BIM.
Next, to determine whether these changes were associated with resistance to glucocorticoid therapy in ALL, the authors examined a number of human ALL patient-derived xenografts (PDX) that had been extensively characterised for glucocorticoid response and annotated as sensitive or resistant (n=5 vs. n=5). They integrated the findings from chromatin immunoprecipitation, accessibility, and gene expression by RNA sequencing to identify gene expression changes that were associated with open chromatin and glucocorticoid receptor binding. A relatively small number of genes were identified that showed increased expression in sensitive ALL PDX, and these correlated with open chromatin and higher H3K27Ac marks (an epigenetic marker for active gene enhancers). The converse was also true, as downregulated genes had fewer H3K27Ac marks and higher levels of DNA methylation. Gene expression analysis linked these genes with apoptotic signalling, but also with B-cell receptor signalling, an integral component of glucocorticoid efficacy in ALL. Again, there was a striking association of induced chromatin accessibility at the proapoptotic BIM locus, and sensitivity to glucocorticoids. These observations were validated functionally using a luciferase reporter assay. Moreover, chromatin confirmation capture was used in the PDX models to directly link CTCF-mediated DNA looping between the BIM promoter and this putative enhancer region. Ablation of this CTCF-binding motif was sufficient to prevent BIM induction after corticosteroids.
In Brief
This article demonstrates that lymphocytes are pre-programmed for glucocorticoid binding that is mediated through a lineage specific chromatin architecture, and that the structural protein CTCF may be involved in manipulating or maintaining this architecture. This work helps explain a recurring clinical observation that we often take for granted (the tissue specific effects of glucocorticoids in lymphoid malignancy) and develops this concept to explain acquired corticosteroid resistance in patients with leukemia. These findings may be used to rationally sequence epigenetic modulators that induce conformational change within chromatin, together with glucocorticoids or chemotherapy, to resensitize treatment-resistant ALL and improve patient outcomes.
References
Competing Interests
Dr. Lane indicated no relevant conflicts of interest.