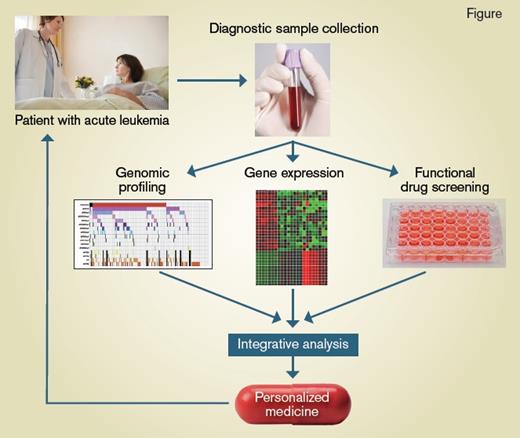
Schematic showing the process from disease sample collection to personalized medicine for treating AML patients
Process from sample collection to personalized medicine for AML patients.
Process from sample collection to personalized medicine for AML patients.
In his ancient military treatise, The Art of War, Sun Tzu teaches the fundamental importance of a deep and detailed understanding of the enemy and the self in order to guarantee success on the battlefield. Despite the many advances in analytical genomics and a rapidly growing pharmacologic armamentarium, acute myeloid leukemia (AML) remains the archetypal enemy of patients and hematologists. Linking this knowledge of the enemy with our own strategies to treat leukemia remains a key and unmet need.
AML is a devastating blood cancer that leads to the death of more than 10,000 patients per year in the United States, and many more internationally. AML is genetically and phenotypically diverse, and this genetic heterogeneity has a major effect on survival after chemotherapy treatment.1 Molecularly targeted therapies have revolutionized the treatment of blood cancers; for example, the routine use of ABL kinase inhibitors in chronic myeloid leukemia, or ATRA plus arsenic in acute promyelocytic leukemia, both lead to long-term survival in more than 90 percent of patients. These initial spectacular successes have overshadowed the modest benefits from newer targeted treatments; however, it follows that specific targeting of oncogenic driver lesions may improve survival across many genetic subtypes of AML. The Beat AML trial, a large collaborative effort spearheaded by the Leukemia and Lymphoma Society, seeks to test this hypothesis in a prospective clinical trial designed to empower clinicians and patients by linking real-time genetic information with molecularly targeted therapeutic options.
In the first published results from the Beat AML Master Trial, Dr. Jeffrey W. Tyner and colleagues took an ambitious approach to integrate clinical, cytogenetic, molecular genetics, and transcriptional analysis, together with in vitro testing of primary samples, examining drug sensitivity against 122 different compounds. Both genetic factors and transcriptional signatures were associated with drug sensitivity. Genetic subgroups, including TP53 or ASXL1 mutations, were associated with widespread resistance in the drug screen, whereas other lesions (e.g., NRAS mutations) showed resistance to many drugs but demonstrated sensitivity to MAPK pathway inhibition.
A major challenge in treating AML has been understanding the complex interactions between co-occurring genetic factors in the disease. Dr. Tyner and colleagues examined patterns of genetic cooperativity or mutual exclusivity and validated these in an independent patient cohort. For example, NPM1c mutation is one of the most frequent genetic alterations in AML, but was not found in PML-RARalpha or CBFbeta-MYH11 AML. In contrast, there was a strong positive association of NPM1c mutation with DNMT3A mutation. Combining these data with the in vitro drug sensitivity screen, the authors identified antileukemic activity of the tyrosine kinase inhibitors ibrutinib and entospletinib in NPM1c-mutant or FLT3ITD-mutant AML. However, this increased sensitivity was not seen in AML with FLT3ITD and concurrent DNMT3A mutations, or sole DNMT3A mutations. The authors hypothesized that this may reflect activity against spleen-associated tyrosine kinase (SYK), though this was not specifically demonstrated.
To understand the mechanism of individual drug responses (Figure), the authors were able to generate unique gene expression signatures from primary AML samples with the highest 20 percent versus lowest 20 percent of responding samples for each drug. This was successful in 78 of the 112 drugs tested (FDR <0.05). Finally, the authors performed integrated, multivariate modelling to extend these findings beyond individual mutational or gene expression changes. Using this approach, they were able to identify co-occurrences of genetic mutations and gene expression clusters that predicted response. It will be fascinating to see whether these biological insights can be translated into meaningful patient responses on the therapeutic arm of the clinical trial.
In Brief
Dr. Tyner and colleagues provide functional annotation of specific genetic AML subtypes using in vitro drug sensitivity screening and a systems approach to integrate cytogenetic, molecular, and gene expression data, mapping a path to clinical translation that will be tested in other aspects of the Beat AML trial. This article builds on the seminal work of Dr. Elli Papaemmanuil and colleagues,2 who had previously used clinical trial data to correlate comprehensive genetic factors with clinical response rates and survival after standard chemotherapy. These two major cohorts have some important differences; for example, the Beat AML cohort comprised an older group of patients (median age, 61 vs. 50 years)2 with fewer de novo AML samples. Importantly, the authors provide powerful online tools to interrogate clinical and genetic features in a portal that is accessible for clinical and research hematologists. Through this provision of publicly available, comprehensively annotated datasets, these results contribute to a global network of big data that can be used for follow-up projects that enables the testing or validation of translational hypotheses and new prognostic information.3
This work reinforces the finding that prognosis in AML is defined by tumour specific factors, and is predominantly specified by response to treatment. Predictive algorithms that enable careful selection of the most appropriate and effective therapies are likely to be of utmost importance to improve survival for patients with this devastating blood cancer.
Note: ASH has partnered with the Leukemia & Lymphoma Society to help spread the word about the pivotal Beat AML trial.
References
Competing Interests
Dr. Lane indicated no relevant conflicts of interest.