Although clinicomorphologic classification of malignancies has been the mainstay of diagnostic pathology for years, the past decade has seen a progressive trend to use molecular genetic data to inform diagnosis and prognosis, and to predict response to a rapidly evolving arsenal of therapies. In fact, hematologic neoplasms have been at the forefront of this paradigm shift that is sweeping through oncology. This deeper genetic understanding has resulted in more precise diagnostic classification (e.g., BCR-ABL1 in chronic myeloid leukemia [CML], KIT mutations in systemic mastocytosis) as well as created subclassifications that stratified patients both based upon prognosis (e.g., IKZF1 deletions in B-cell precursor lymphoblastic leukemia and TP53 mutations in acute myeloid leukemia [AML]) and targeted therapeutic opportunities (e.g., IDH1/2 and FLT3 mutations in AML, BCR-ABL1 rearrangements in CML and ALL). The year 2018 saw the field undergo a new paradigm shift. Rather than refining existing clinicomorphologic categories, new molecularly defined categories have been identified in the myeloproliferative neoplasms (MPNs) that transcend the traditional classification scheme and provide a new approach to diagnosis and prognostication of these diseases.
MPNs are a heterogeneous group of clonal hematopoietic disorders unified by increased numbers of differentiated blood cells. The Philadelphia chromosome (Ph) –negative MPNs include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). The differentiation between these diagnoses has been largely based on clinical parameters such as blood cell counts, and bone marrow morphology. However, there can be substantial clinical and morphologic overlap between these entities, which can create clinical uncertainty due to the wide range of prognoses between and within these classifications. Currently, disease-specific risk-stratification models place patients into broad risk categories that are primarily based on cell counts, age, thrombosis risk, and limited genetic data.1 The latter include driver mutations in JAK2, CALR, and MPL that can be identified in the majority of Ph-negative MPNs. However, these mutations lead to functionally convergent oncogenic JAK-STAT signaling and do not discriminate between the MPNs.
Eight genomic categories of myeloproliferative neoplasms (MPNs): classification schematic, distribution with MPN subtypes, outcomes in chronic disease (polycythemia vera [PV] or essential thrombocythemia [ET]), and outcomes in myelofibrosis (either primary myelofibrosis or post-PV/ET myelofibrosis [MF]). (Reprinted from Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416-1430.)
Eight genomic categories of myeloproliferative neoplasms (MPNs): classification schematic, distribution with MPN subtypes, outcomes in chronic disease (polycythemia vera [PV] or essential thrombocythemia [ET]), and outcomes in myelofibrosis (either primary myelofibrosis or post-PV/ET myelofibrosis [MF]). (Reprinted from Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416-1430.)
Nevertheless, a number of other diverse pathogenic mutations that can influence phenotype and clinical outcome can also be identified in these diseases.2 Our rapidly evolving genetic understanding of MPNs raises the tantalizing possibility of building a more holistic multiparametric model incorporating high-dimensional phenotypic and genomic data that may have predictive power across all traditional categories of Ph-negative MPNs. To address this possibility, Dr. Jacob Grinfeld and colleagues undertook targeted next generation sequencing of exonic regions of 69 myeloid cancer genes to detect single nucleotide variants and copy number changes in a retrospective cohort of 2,035 patients with Ph-negative MPNs — the largest molecularly characterized cohort of patients with MPN.3 Thirty-three recurrent mutated genes were identified with JAK2, CALR, and MPL being the sole somatic variant in a large subset of cases (45%). Using a combination of Bayesian network analysis and Dirichlet process mixture modeling approaches, eight genomic subgroups were identified, each with distinct clinicopathologic phenotypes including outcomes (Figure 1). Importantly, the eight subgroups in many cases contained representation from all three main subtypes of Ph-negative MPNs, consistent with reshuffling of traditional diagnostic categories into these molecularly-defined diagnostic groups. The reconfigured groups demonstrate several recurrent principles that hold true across all myeloid neoplasms.4 For example, the subgroups with TP53 mutations and mutations involving chromatin modifying/RNA splicing genes generally occurred in older patients with poor outcomes (relative to the JAK2 heterozygous subgroup), regardless of MPN phenotype, a pattern seen in other hematologic malignancies.4
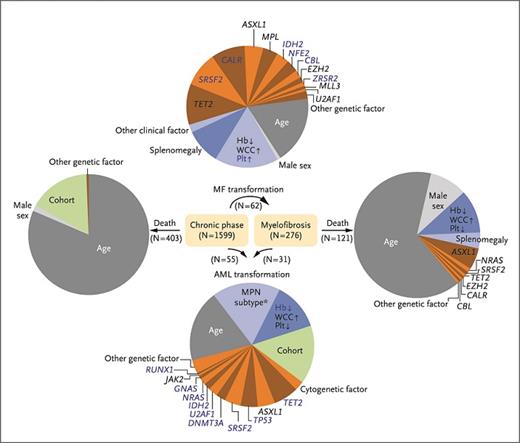
Relative contributions of parameters that contribute to progression to myelofibrosis (MF), acute myeloid leukemia (AML), and death. (Reprinted from Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416-1430.)
Relative contributions of parameters that contribute to progression to myelofibrosis (MF), acute myeloid leukemia (AML), and death. (Reprinted from Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416-1430.)
Furthermore, the authors generated a multiparameter multistate prognostic model, combining 63 clinical and genomic variables to generate predictions for an individual patient across all Ph-negative MPNs (access the MPN personalized risk calculatorcalculator). The model showed high predictive accuracy in internal cross-validation of a training cohort and in an independent external cohort. The resultant prognostic model consistently outperformed the International Prognostic Scoring System (IPSS), the Dynamic IPSS (D-IPSS), as well as high molecular risk category for myelofibrosis and the International Prognostic Score for Essential Thrombocythemia (IPSET). Again, clinical and molecular data predominated the prediction algorithms for progression to myelofibrosis, AML, and death (Figure 2), while traditional morphologic subtype did not. For example, age was the single most powerful predictor of death, while it was relatively less important in predicting progression to AML or myelofibrosis. Conversely, cell counts and genetic factors played a dominant role in predicting the latter. By contrast, the MPN subtype provided only a modest contribution to predicting progression to AML and did not significantly contribute to other state transitions. While retaining the distinction between chronic-phase disease such as ET and PV and myelofibrosis (including both PMF and post-PV/ET myelofibrosis) improved predictive accuracy of the model, the distinction between ET and PV did not improve accuracy. These findings collectively suggest that, at least for multiparameter prognostication using clinical and genomic data, the distinction of certain traditional diagnostic categories (ET vs. PV) may be dispensable.
This study raises the philosophical question regarding the future utility of the conventional classification system of MPNs, and indeed all neoplasms, that relies upon morphologic features with only limited utilization of genetic data. In the past year, the primacy of traditional diagnostic categories has also been challenged in the lymphomas. The results of genomic characterization of large cohorts of diffuse large B cell lymphomas (DLBCL) were published consecutively by two separate groups (also reviewed in the September/October 2018 issue of The Hematologist this year by Drs. Caron Jacobson and Andrew Roberts). Like MPNs, DLBCLs are a heterogeneous group of tumors with variable clinical behavior that may be partially predicted using conventional prognostic models. Dr. Bjoern Chapuy and colleagues5 as well as Dr. Roland Schmitz and colleagues6 described distinct molecular-genetic subtypes of DLBCL that were distributed across conventional germinal center-like (GCB) and activated B cell-like (ABC) subgroups but provided prognostic information independent of Cell-of-Origin and IPI classification. Analogous to MPNs, both these studies offer proof-of-principle that genomic profiling can offer orthogonal prognostic information.
The identification of molecularly defined diagnostic categories that transcend traditional classification schemes in no way suggests that clinical and morphologic information is not needed. However, the findings of the past year have emphasized the added value of a much broader multiparametric approach that synthesizes clinical, morphologic, and extensive molecular genetic data (including somatic, copy number, and structural variants) in support of clinical decision-making. The dimensionality of the data is so vast that simple algorithms will no longer be sufficiently granular. Online “calculators,” such as that provided by Dr. Jacob Grinfeld and colleagues, will be required to generate patient-specific diagnoses. So, while 2018 has been the year of broad molecular diagnostic data, let 2019 usher in the exciting and challenging task of translating these rapidly evolving insights into robust ancillary tools to enrich traditional diagnostic categories and fulfill the promise of precision medicine across all hematologic neoplasms.
References
Competing Interests
Dr. Shanmugam and Dr. Kim indicated no relevant conflicts of interest.

![Eight genomic categories of myeloproliferative neoplasms (MPNs): classification schematic, distribution with MPN subtypes, outcomes in chronic disease (polycythemia vera [PV] or essential thrombocythemia [ET]), and outcomes in myelofibrosis (either primary myelofibrosis or post-PV/ET myelofibrosis [MF]). (Reprinted from Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N Engl J Med. 2018;379:1416-1430.)](https://ash.silverchair-cdn.com/ash/content_public/journal/thehematologist/16/1/10.1182_hem.v16.1.9295/2/m_d395fa1a-3b4d-4299-ad26-7d5735be02f8.jpeg?Expires=1767724573&Signature=RWgKPphjtLV0aYySBbjk5e5A2hUgbefjvdh2YYl-BnB44JkgaXxS5eDDun~vyiTdlIZ~NXKbJKUIQBNHl7wjH1j~GF75O75-BcY7LlhsZw-xKLvvIqKOIbaoYjT15-mTBEJ3aoe8Fs7LUOXHATwbNm3GdPf5LOoZH8uPwpn3MIifv47LbkXjFcj7xGBNjnjZQGJg2iMZ604TQDZCQOIHtxq1RTRk2DiYFSrnVq4EVb7U-U0tXufPNABMWNXHKF43RdjyCAMTKT-k5yvss-XhirdfKLlHHBADhGTwvZ8w6yyfSVNRluayYmgCNg-WKgTr3ot2kx91ge~EH97fM3TPBw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)